Biology:Community fingerprinting
Community fingerprinting is a set of molecular biology techniques that can be used to quickly profile the diversity of a microbial community. Rather than directly identifying or counting individual cells in an environmental sample, these techniques show how many variants of a gene are present. In general, it is assumed that each different gene variant represents a different type of microbe. Community fingerprinting is used by microbiologists studying a variety of microbial systems (e.g. marine, freshwater, soil, and human microbial communities) to measure biodiversity or track changes in community structure over time. The method analyzes environmental samples by assaying genomic DNA. This approach offers an alternative to microbial culturing, which is important because most microbes cannot be cultured in the laboratory.[1] Community fingerprinting does not result in identification of individual microbe species; instead, it presents an overall picture of a microbial community. These methods are now largely being replaced by high throughput sequencing, such as targeted microbiome analysis (e.g., 16s rRNA sequencing) and metagenomics.
Use
A fingerprinting analysis begins with an environmental sample (e.g. seawater or soil), from which total DNA is extracted. (Total DNA contains a mix of genetic material from all the microbes present in the sample.) A particular gene or DNA region is then selected as a target for analysis, under the assumption that each microbe species will have a different gene variant (also called a "phylotype"). Different methods (see below) can be used to visualize the phylotypes present in a sample. Because the aim of community fingerprinting is to gain an overall understanding of community structure, it is a particularly useful technique for analyzing time-series data collected from the field. For example, one could study the pattern of microbial succession in a habitat, or one could examine the response of a microbial community to an environmental perturbation, such as the release of a pollutant. Depending on what information is desired, different genes may be targeted. The most common are small subunit ribosomal RNA (rRNA) genes, such as 16S rRNA. These genes are frequently used in microbial phylogenetic analyses, so well-established techniques exist for their study. Other genes of interest might be those that are key in various metabolic processes.[1]
Advantages and disadvantages
The advantages of community fingerprinting are that it can be performed quickly and relatively cheaply, and the analyses can accommodate a large number of samples simultaneously.[2][3] These properties make community fingerprinting especially useful for monitoring changes in microbial communities over time. Also, fingerprinting techniques do not require one to have a priori sequence data for organisms in a sample. A disadvantage of community fingerprinting is that it results in largely qualitative, not quantitative data.[4] When using qualitative data, it can be difficult to compare patterns observed in different studies or between different investigators. Also, community fingerprinting does not directly identify taxa in an environmental sample, though the data output from certain techniques (e.g. DGGE) can be analyzed further if one desires identification. Some authors point to poor reproducibility of results for certain fingerprinting methods,[3] while other authors have criticized the inaccuracy of abundance estimates and the inability of some techniques to capture the presence of rare taxa.[5] For example, it is difficult for the DGGE method to detect microbes that comprise less than 0.5%-1% of a bacterial community.[6]
Techniques
This section presents three methods of community fingerprinting.
Terminal restriction fragment length polymorphism (T-RFLP)
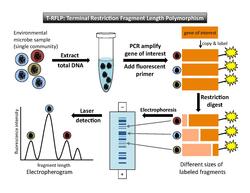
Terminal restriction fragment length polymorphism (T-RFLP) is a method that uses fluorescently-labeled DNA fragments to produce a community fingerprint.[2][7] This section presents a brief explanation of T-RFLP in the specific context of community fingerprinting. For a more detailed explanation, refer to the T-RFLP article.
Procedure
To perform T-RFLP (Figure 1), one must select a target gene (e.g. the 16S rRNA gene) to amplify by PCR. At least one primer used in the PCR reaction is fluorescently labeled at the 5´ end. After PCR amplification, each copied DNA segment carries the fluorescent label. Next, restriction enzymes are used to cut the amplified DNA at specific recognition sites. The underlying assumption is that each microbe in the sample will have a different sequence on the target gene, so a restriction enzyme will cut each microbe's DNA in a different place. (Each different restriction site is considered to represent a single operational taxonomic unit [OTU]). Thus, the enzyme will produce one fragment length for each type of microbe present in the sample. The result of digestion is a set of restriction fragments of different lengths, each of which is fluorescently labeled at one end. These are known as "terminal fragments" because they are labeled at the end where the PCR primer attached. (The unlabeled ends are not recorded in the final analysis.) Next, the fragments are separated by size through either gel or capillary electrophoresis. Laser detection captures the size and fluorescence-intensity patterns of the terminal fragments. DNA standards of known size and fluorescence are included in the analysis as references. By setting a minimum threshold for fluorescence, background noise will be excluded.[4]
Data output and interpretation
The output of the laser detection step is an electropherogram that shows a series of peaks, with fragment length on the horizontal axis and fluorescence intensity on the vertical axis (Figure 1). A second output[4] is a data table that lists the migration time of the fragments, the size in base pairs of each peak, and the height of and area under each peak.
The theoretical basis of T-RFLP assumes that peaks at different positions along the horizontal axis represent different types of organisms (or OTUs). The area under each fluorescence intensity peak is a proxy for relative abundance of each phylotype in the community. However, a number of caveats must be taken into account. Different types of organisms may share a restriction site in the gene of interest; if that is the case, these organisms would not be distinguished as different peaks on the electropherogram. Furthermore, area under a peak represents relative abundance rather than absolute abundance, and there are biases in abundance measurement and PCR amplification. For example, organisms that are scarce in the original total DNA sample will not be amplified enough to be detected in the final analysis.[7] This leads to underestimation of community diversity. Liu et al.[7] cite other possible factors that may distort results, including "differences in gene copy number between species and biases introduced during cell lysis, DNA extraction, and PCR amplification" (p. 4521). For those who seek detailed technical information, Marsh[2] provides a catalog of potential biases that could be introduced in each step of the T-RFLP process.
Advantages and disadvantages
The major advantages of T-RFLP are that it is fast and can easily accommodate many samples.[2] Also, the visual output simplifies comparison of community structure patterns across different samples. A disadvantage is the broad, qualitative nature of the data output, which must be interpreted with the above caveats in mind. Also, direct identification of microbes in a sample is not possible through T-RFLP.
Applications
Disayathanoowat et al.[8] used T-RFLP to assess the microbial gut community, or microbiome, in two species of honeybees in Thailand. They found that the two species harbor different microbial communities and that the microbiome changes over the lifetime of the bees.
Joo et al.[9] tested T-RFLP as a method for phytoplankton monitoring. The authors collected environmental water samples from reservoirs in a time series. After comparison of samples with known terminal restriction fragments (from a database built from cultures), they concluded that T-RFLP can be used effectively as a technique for monitoring changes in the phytoplankton community. However, diversity and abundance estimates were found to be less accurate than those found through other methods.
Denaturing gradient gel electrophoresis (DGGE)
Procedure
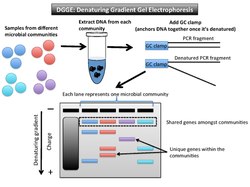
Denaturing gradient gel electrophoresis (DGGE) is a microbial fingerprinting technique that separates amplicons of roughly the same size based on sequence properties[1] (Figure 2). These properties dictate the threshold at which DNA denatures. The DGGE gel uses a gradient DNA denaturant (a mixture of urea and formamide), or a linear temperature gradient.[10] When the fragment reaches its melting point (threshold of enough denaturant), it stops moving. This is due to the fact that a partially melted double-stranded DNA can no longer migrate through the gel.[11] A GC clamp (about 40 bases with high GC content) is used as a special primer to anchor the PCR fragments together once they have denatured.[12]
Data output and interpretation
Each lane on a gel represents one microbial community. The shared bands among the samples are the same size and roughly the same position on the gel. The gene variants that are not shared amongst microbial community samples do not match up horizontally with others. For example, if the gene of interest is 16S rRNA, as it was when the technique was first described, the PCR-amplified fragments will be in the same vertical location because they are all roughly the same size.[13] Another target gene may have greater variation in length, but the denaturant gradient uses a second element (of melting point) to further distinguish between the samples. The DGGE gel will separate genes of the same size based on base sequence.
This technique shows to what extent microbial communities are the same or different in taxonomic composition. Each band in a different location on the gel represents a different phylotype (one unique sequence of a phylogenetic marker gene).[1] For microbial communities this method profiles many individual 16S rRNA sequences.[14] The number of bands at differing horizontal positions can be used to estimate the level of biodiversity in that sample and infer phylogenetic affiliation.[10] In order to know more about phylogenetic affiliation, one could excise those bands from the gel and then sequence them.
Advantages and disadvantages
The use of denaturing profile serves as a way to separate DNA fragments of similar sizes. This is beneficial in assessing microbial diversity due to the fact that the 16S rRNA gene does not vary much in size across bacterial phyla. The DGGE gel provides a quick way of looking at biodiversity in a microbial sample and does not preclude the option of sequencing the bands of interest. This method does not require that the microbes be cultured in the lab and does not require any sequence data needed to design probes for hybridization methods.[10] The main disadvantage is that this is a qualitative assessment of biodiversity and one must sequence the genes in order to make inferences about the phylogenetic relatedness. Another disadvantage is that the GC clamp can be variable each time it is synthesized. This leads to the potential for different DGGE profiles for the same 16S rRNA sequence.[12]
Applications
Stephen et al.[15] utilized DGGE for a rapid analysis of Proteobacteria in soil. They obtained an initial assessment of microbial diversity in their environmental samples from soil maintained for 36 years at the various pH values. They combined DGGE and hybridization techniques by probing the DNA fragments to obtain more detail about the natural populations. In this study, they were looking at a group of closely related bacterial types, all autotrophic β–proteobacterial ammonia oxidizers. The 16S rDNA samples yielded ambiguous overlapping bands when run out on a gel. The ambiguous overlapping bands were separated with cluster-specific radiolabelled probes, which yielded information of the relative abundance of the different genotypes in samples.
Ward et al.[14] examined cyanobacterial mat communities in a Yellowstone hot spring by using DGGE analysis of 16S rRNA gene segments of aerobic chemoorganotrophic populations. DGGE allowed them to profile the community gaining new insight to diversity. They characterized the bands of interest by purifying and sequencing and detected many lineages previously unknown to be present.
(Automated) ribosomal intergenic spacer analysis (ARISA)
Procedure
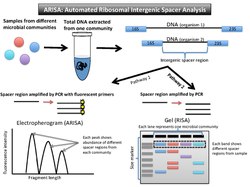
(Automated) ribosomal intergenic spacer analysis, or (A)RISA (Figure 3), takes advantage of the fact that prokaryotic DNA encodes for two highly conserved genes, the 16SrRNA gene and the 23SrRNA gene. These encode the small and large subunit genes in the rRNA operon. Between these two genes, there is an internal transcribed spacer (ITS) region. Due to the fact it is non-coding for proteins, it is a highly variable nucleotide sequence and length. Once DNA is isolated from a community, PCR amplifies this spacer region. The fragments can be run out on a gel (RISA), or the fluorescent primers can be translated into peaks in abundance of the different fragments lengths on an electropherogram (ARISA).[16][17]
Data output and interpretation
Due to variable non-coding regions, the output for RISA is a gel with different banding patterns, and output for ARISA is an electropherogram with different peaks (similar to T-RFLP).
The brightness of the fluorescently labeled primers correlates to how prevalent that bacterial type is in the community. The banding pattern on the gel can be interpreted as a community-specific profile. Each DNA band or peak indicates at least one representative of that organism.[16] In RISA, the bands on gel that do not match up in length represent different organisms in the community because they have different spacer regions between the two highly conserved genes. The electropherogram shows peaks correlating to the relative abundance of that spacer region in the sample.
Advantages and disadvantages
ARISA can have a higher resolution in detecting microbial diversity as compared to T-RFLP.[18] This fingerprinting method is a quick and sensitive method to estimate microbial diversity. The observed length heterogeneities can be compared to databases for overlap with culturable organisms.[16] One can design phylum-level oligonucleotide primers to get at questions regarding phylogenetic groups.[16] One disadvantage to ARISA is the fact that a single organism may contribute more than one peak to the community profile. Unrelated organisms can also have similar spacer lengths, which leads to underestimates of community diversity. Due to these biases, researchers often use this method on multiple samples from each community in order to get an average assessment.
Applications
Ranjard et al.[17] discuss several examples of types of studies in which RISA can be used. They cite several studies that have used this technique to fingerprint bacterial communities following such disturbances as antibiotic treatment, mercury stress and deforestation. They also demonstrated the successful use of ARISA for characterizing fungal communities, which is an aspect of microbial ecology that remains to be fully explored.
Schloss et al.[19] conducted a study examining environmental variables and linking them to changes in the microbial ecology of a compost pile. They used ARISA to profile the community structure and look at microbial succession over stages in the composting process. They took DNA samples and sequenced the 16S rRNA gene to identify community members at the different phases of the process. Then they used ARISA to look at community-wide changes and then matched up the 16S rRNA gene sequence with the most common ARISA fragments to identify these microbial community members.
References
- ↑ 1.0 1.1 1.2 1.3 Madigan, M.T.; J.M. Martinko; P.V. Dunlap; D.P. Clark (2009). Brock Biology of Microorganisms (12th ed.). San Francisco, CA: Pearson Education Inc..
- ↑ 2.0 2.1 2.2 2.3 Marsh, T.L. (2005). "Culture‐Independent Microbial Community Analysis with Terminal Restriction Fragment Length Polymorphism". Culture-independent microbial community analysis with terminal restriction fragment length polymorphism. Methods in Enzymology. 397. pp. 308–329. doi:10.1016/s0076-6879(05)97018-3. ISBN 9780121828028.
- ↑ 3.0 3.1 Grant, A.; L.A. Ogilvie (2004). "Name that microbe: rapid identification of taxa responsible for individual fragments in fingerprints of microbial community structure". Molecular Ecology Notes 4 (1): 133–136. doi:10.1111/j.1471-8286.2004.00590.x.
- ↑ 4.0 4.1 4.2 Osborn, A.M.; E.R.B. Moore; K.N. Timmis (2000). "An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics". Environmental Microbiology 2 (1): 39–50. doi:10.1046/j.1462-2920.2000.00081.x. PMID 11243261.
- ↑ Bent, S.J.; J.D. Pierson; L.J. Forney (2007). "Measuring species richness based on microbial community fingerprints: the emperor has no clothes". Applied and Environmental Microbiology 73 (7): 2399–2401. doi:10.1128/aem.02383-06. PMID 17403942. Bibcode: 2007ApEnM..73.2399B.
- ↑ Kan, J.; K. Wang; F. Chen (2006). "Temporal variation and detection limit of an estuarine bacterioplankton community analyzed by denaturing gradient gel electrophoresis (DGGE)". Aquatic Microbial Ecology 42: 7–18. doi:10.3354/ame042007.
- ↑ 7.0 7.1 7.2 Liu, W.-T.; T. L. Marsh; H. Cheng; L.J. Forney (1997). "Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA". Applied and Environmental Microbiology 63 (11): 4517–4522. doi:10.1128/AEM.63.11.4516-4522.1997. PMID 9361437. Bibcode: 1997ApEnM..63.4516L.
- ↑ Disayathanoowat, T.; J.P.W. Young; T. Helgason; P. Chantawannakul (2012). "T-RFLP analysis of bacterial communities in the midguts of Apis mellifera and Apis cerana honey bees in Thailand". FEMS Microbiology Ecology 79 (2): 273–281. doi:10.1111/j.1574-6941.2011.01216.x. PMID 22092273.
- ↑ Joo, S.; S.-R. Lee; S. Park (2010). "Monitoring of phytoplankton community structure using terminal restriction fragment length polymorphism (T-RFLP)". Journal of Microbiological Methods 81 (1): 61–68. doi:10.1016/j.mimet.2010.01.025. PMID 20138925.
- ↑ 10.0 10.1 10.2 Muyzer, G. (1999). "DGGE/TGGE a method for identifying genes from natural ecosystems". Current Opinion in Microbiology 2 (3): 317–322. doi:10.1016/s1369-5274(99)80055-1. PMID 10383868.
- ↑ Fischer, S.G.; L.S. Lerman (1983). "DNA fragments differing by single base-pair substitutions are separated in denaturing electrophoreseis analysis of polymerase chain reaction-amplified genes coding for 16S rRN". PNAS 80 (6): 1579–1583. doi:10.1073/pnas.80.6.1579. PMID 6220406.
- ↑ 12.0 12.1 Rettedal, E.A.; S. Clay; V. S. Brözel (2010). "GC-clamp primer batches yield 16S rRNA gene amplicon pools with variable GC clamps, affecting denaturing gradient gel electrophoresis profiles". FEMS Microbiology Letters 312 (1): 55–62. doi:10.1111/j.1574-6968.2010.02097.x. PMID 20831594.
- ↑ Muyzer, G.; E.C. De Waal; A.G. Uitterlinden (1993). "Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRN". Applied and Environmental Microbiology 59 (3): 695–700. doi:10.1128/AEM.59.3.695-700.1993. PMID 7683183. Bibcode: 1993ApEnM..59..695M.
- ↑ 14.0 14.1 Ward, D.M.; M. J. Ferris; S. C. Nold; M. M. Bateson (1998). "A natural view of microbial biodiversity within hot spring cyanobacterial mat communities". Microbiology and Molecular Biology Reviews 62 (4): 1353–1370. doi:10.1128/MMBR.62.4.1353-1370.1998. PMID 9841675.
- ↑ Stephen, J.R.; G. A. Kowalchuk; M-A. V. Bruns; A. E. McCaig; C. J. Phillips; T. M. Embley; J. I. Prosse (1998). "Analysis of β-subgroup Proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing". Applied and Environmental Microbiology 64 (8): 2958–2965. doi:10.1128/AEM.64.8.2958-2965.1998. PMID 9687457. Bibcode: 1998ApEnM..64.2958S.
- ↑ 16.0 16.1 16.2 16.3 Fisher, M.M.; E.W. Triplett (1999). "Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities". Applied and Environmental Microbiology 65 (10): 4630–4636. doi:10.1128/AEM.65.10.4630-4636.1999. PMID 10508099. Bibcode: 1999ApEnM..65.4630F.
- ↑ 17.0 17.1 Ranjard, L.; F. Poly; J.-C. Lata; C. Mougel; J. Thioulouse; S. Nazaret (2001). "Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability". Applied and Environmental Microbiology 67 (10): 4479–4487. doi:10.1128/aem.67.10.4479-4487.2001. PMID 11571146. Bibcode: 2001ApEnM..67.4479R.
- ↑ Danovaro, R.; G. M. Luna; A. Dell’Anno; B. Pietrangeli (2006). "Comparison of two fingerprinting techniques, terminal restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis, for determination of bacterial diversity in aquatic environments". Applied and Environmental Microbiology 72 (9): 5982–5989. doi:10.1128/aem.01361-06. PMID 16957219. Bibcode: 2006ApEnM..72.5982D.
- ↑ Schloss, P.D.; A.G. Hay; D.B. Wilson; L.P. Walker (2003). "Tracking temporal changes in bacterial community fingerprints during the initial stages of compostin". FEMS Microbiology Ecology 46 (1): 1–9. doi:10.1016/s0168-6496(03)00153-3. PMID 19719577.
 |