Biology:Transcriptional amplification
In genetics, transcriptional amplification is the process in which the total amount of messenger RNA (mRNA) molecules from expressed genes is increased during disease, development, or in response to stimuli.
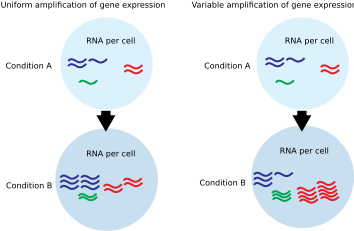
In eukaryotic cells, the transcribing activity of RNA Polymerase II results in mRNA production. Transcriptional amplification is specifically defined as the increase in per-cell abundance of this set of expressed mRNAs. Transcriptional amplification is caused by changes in the amount or activity of transcription-regulating proteins.
Mechanisms of transcriptional amplification
Gene expression is regulated by numerous types of proteins that directly or indirectly influence transcription by RNA Polymerase II. As opposed to transcriptional activators or repressors that selectively activate or repress specific genes, amplifiers of transcription act globally on expressed genes.
Several known regulators of transcriptional amplification have been characterized including the oncogene Myc,[1][2] the Rett syndrome protein MECP2,[3] and the BET bromodomain protein BRD4.[4] In particular, the Myc protein amplifies transcription by binding to promoters and enhancers of active genes where it directly recruits the transcription elongation factor P-TEFb. Furthermore, the BRD4 protein is a regulator of Myc activity.
Identifying and measuring transcriptional amplification
Commonly used gene expression experiments interrogate the expression of one gene (qPCR) or many genes (microarray, RNA-Seq). These techniques generally measure relative mRNA levels and employ normalization methods that assume only a small number of genes show altered expression.[5] In contrast, single cell or cell-count normalized absolute measurements of mRNA abundance are required to reveal transcriptional amplification.[6] Additionally, global measurements of mRNA or total mRNA per cell can also uncover evidence for transcriptional amplification.[7][8]
Cells in which transcription has been amplified have additional hallmarks suggesting that amplification has occurred. Cells with increased mRNA levels may be larger, consistent with an increased abundance of gene products. This increase in the amount of gene products may result in a decreased doubling time.
Role in disease
Transcriptional amplification has been implicated in cancer,[9][10] Rett syndrome,[11] heart disease,[12] Down syndrome,[13] and cellular aging.[14] In cancer, Myc-driven transcriptional amplification is posited to help tumor cells overcome rate-limiting constraints in growth and proliferation.[15] Drugs that target the transcription or mRNA processing machinery are known to be particularly effective against Myc-driven tumor models,[16][17] suggesting that dampening of transcriptional amplification can have anti-tumor effects. Similarly, small molecules targeting the BET bromodomain protein BRD4, which is up-regulated during heart failure, can block cardiac hypertrophy in mouse models.[18][19] In Rett syndrome, which is caused by loss of function of the transcriptional regulator MeCP2, MeCP2 was shown to specifically amplify transcription in neurons and not neuronal precursors.[20] Restoration of MeCP2 reverses disease symptoms associated with Rett syndrome[21][22]
References
- ↑ Lin, CY; Lovén, J; Rahl, PB; Paranal, RM; Burge, CB; Bradner, JE; Lee, TI; Young, RA (28 September 2012). "Transcriptional amplification in tumor cells with elevated c-Myc.". Cell 151 (1): 56–67. doi:10.1016/j.cell.2012.08.026. PMID 23021215.
- ↑ Nie, Z; Hu, G; Wei, G; Cui, K; Yamane, A; Resch, W; Wang, R; Green, DR et al. (28 September 2012). "c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells.". Cell 151 (1): 68–79. doi:10.1016/j.cell.2012.08.033. PMID 23021216.
- ↑ Li, Y; Wang, H; Muffat, J; Cheng, AW; Orlando, DA; Lovén, J; Kwok, SM; Feldman, DA et al. (3 October 2013). "Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons.". Cell Stem Cell 13 (4): 446–58. doi:10.1016/j.stem.2013.09.001. PMID 24094325.
- ↑ Anand, P; Brown, JD; Lin, CY; Qi, J; Zhang, R; Artero, PC; Alaiti, MA; Bullard, J et al. (1 August 2013). "BET bromodomains mediate transcriptional pause release in heart failure.". Cell 154 (3): 569–82. doi:10.1016/j.cell.2013.07.013. PMID 23911322.
- ↑ Hannah, MA; Redestig, H; Leisse, A; Willmitzer, L (July 2008). "Global mRNA changes in microarray experiments.". Nature Biotechnology 26 (7): 741–2. doi:10.1038/nbt0708-741. PMID 18612292.
- ↑ Lovén, J; Orlando, DA; Sigova, AA; Lin, CY; Rahl, PB; Burge, CB; Levens, DL; Lee, TI et al. (26 October 2012). "Revisiting global gene expression analysis.". Cell 151 (3): 476–82. doi:10.1016/j.cell.2012.10.012. PMID 23101621.
- ↑ Lin, CY; Lovén, J; Rahl, PB; Paranal, RM; Burge, CB; Bradner, JE; Lee, TI; Young, RA (28 September 2012). "Transcriptional amplification in tumor cells with elevated c-Myc.". Cell 151 (1): 56–67. doi:10.1016/j.cell.2012.08.026. PMID 23021215.
- ↑ Kanno, J; Aisaki, K; Igarashi, K; Nakatsu, N; Ono, A; Kodama, Y; Nagao, T (29 March 2006). ""Per cell" normalization method for mRNA measurement by quantitative PCR and microarrays.". BMC Genomics 7: 64. doi:10.1186/1471-2164-7-64. PMID 16571132.
- ↑ Lin, CY; Lovén, J; Rahl, PB; Paranal, RM; Burge, CB; Bradner, JE; Lee, TI; Young, RA (28 September 2012). "Transcriptional amplification in tumor cells with elevated c-Myc.". Cell 151 (1): 56–67. doi:10.1016/j.cell.2012.08.026. PMID 23021215.
- ↑ Nie, Z; Hu, G; Wei, G; Cui, K; Yamane, A; Resch, W; Wang, R; Green, DR et al. (28 September 2012). "c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells.". Cell 151 (1): 68–79. doi:10.1016/j.cell.2012.08.033. PMID 23021216.
- ↑ Li, Y; Wang, H; Muffat, J; Cheng, AW; Orlando, DA; Lovén, J; Kwok, SM; Feldman, DA et al. (3 October 2013). "Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons.". Cell Stem Cell 13 (4): 446–58. doi:10.1016/j.stem.2013.09.001. PMID 24094325.
- ↑ Anand, P; Brown, JD; Lin, CY; Qi, J; Zhang, R; Artero, PC; Alaiti, MA; Bullard, J et al. (1 August 2013). "BET bromodomains mediate transcriptional pause release in heart failure.". Cell 154 (3): 569–82. doi:10.1016/j.cell.2013.07.013. PMID 23911322.
- ↑ Lane, AA; Chapuy, B; Lin, CY; Tivey, T; Li, H; Townsend, EC; van Bodegom, D; Day, TA et al. (June 2014). "Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation.". Nature Genetics 46 (6): 618–23. doi:10.1038/ng.2949. PMID 24747640.
- ↑ Hu, Z; Chen, K; Xia, Z; Chavez, M; Pal, S; Seol, JH; Chen, CC; Li, W et al. (15 February 2014). "Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging.". Genes & Development 28 (4): 396–408. doi:10.1101/gad.233221.113. PMID 24532716.
- ↑ Ruggero, D (1 December 2009). "The role of Myc-induced protein synthesis in cancer.". Cancer Research 69 (23): 8839–43. doi:10.1158/0008-5472.CAN-09-1970. PMID 19934336.
- ↑ Christensen, CL; Kwiatkowski, N; Abraham, BJ; Carretero, J; Al-Shahrour, F; Zhang, T; Chipumuro, E; Herter-Sprie, GS et al. (8 December 2014). "Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor.". Cancer Cell 26 (6): 909–22. doi:10.1016/j.ccell.2014.10.019. PMID 25490451.
- ↑ Hsu, TY; Simon, LM; Neill, NJ; Marcotte, R; Sayad, A; Bland, CS; Echeverria, GV; Sun, T et al. (17 September 2015). "The spliceosome is a therapeutic vulnerability in MYC-driven cancer.". Nature 525 (7569): 384–8. doi:10.1038/nature14985. PMID 26331541. Bibcode: 2015Natur.525..384H.
- ↑ Anand, P; Brown, JD; Lin, CY; Qi, J; Zhang, R; Artero, PC; Alaiti, MA; Bullard, J et al. (1 August 2013). "BET bromodomains mediate transcriptional pause release in heart failure.". Cell 154 (3): 569–82. doi:10.1016/j.cell.2013.07.013. PMID 23911322.
- ↑ Stratton, MS; Lin, CY; Anand, P; Tatman, PD; Ferguson, BS; Wickers, ST; Ambardekar, AV; Sucharov, CC et al. (2 August 2016). "Signal-Dependent Recruitment of BRD4 to Cardiomyocyte Super-Enhancers Is Suppressed by a MicroRNA.". Cell Reports 16 (5): 1366–78. doi:10.1016/j.celrep.2016.06.074. PMID 27425608.
- ↑ Li, Y; Wang, H; Muffat, J; Cheng, AW; Orlando, DA; Lovén, J; Kwok, SM; Feldman, DA et al. (3 October 2013). "Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons.". Cell Stem Cell 13 (4): 446–58. doi:10.1016/j.stem.2013.09.001. PMID 24094325.
- ↑ Luikenhuis, S; Giacometti, E; Beard, CF; Jaenisch, R (20 April 2004). "Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice.". Proceedings of the National Academy of Sciences of the United States of America 101 (16): 6033–8. doi:10.1073/pnas.0401626101. PMID 15069197. Bibcode: 2004PNAS..101.6033L.
- ↑ Garg, SK; Lioy, DT; Cheval, H; McGann, JC; Bissonnette, JM; Murtha, MJ; Foust, KD; Kaspar, BK et al. (21 August 2013). "Systemic delivery of MeCP2 rescues behavioral and cellular deficits in female mouse models of Rett syndrome.". The Journal of Neuroscience 33 (34): 13612–20. doi:10.1523/JNEUROSCI.1854-13.2013. PMID 23966684.
 |