Biology:Dictyoglomi-1 RNA motif
| Dictyoglomi-1 RNA | |
|---|---|
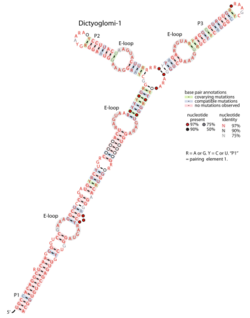 Consensus secondary structure of Dictyoglomi-1 RNAs. This figure is adapted from a previous publication.[1] | |
| Identifiers | |
| Symbol | Dictyoglomi-1 |
| Rfam | RF01703 |
| Other data | |
| RNA type | Gene; sRNA; |
| Domain(s) | Bacteria; |
| SO | 0000655 |
| PDB structures | PDBe |
The Dictyoglomi-1 RNA motif (also called dct-1) is a conserved RNA structure that was discovered via bioinformatics.[1][2] Only four instances of the RNA were detected, and all are in the bacterial phylum Dictyoglomota (formerly Dictyoglomi), whose members have not been extensively studied. The RNA might have a cis-regulatory role, but the evidence is ambiguous. Because of the few instances of Dictyoglomi-1 RNAs known, it is also unknown whether the RNA structure might extend further in the 5′ or 3′ direction, or in both directions.
The Dictyoglomi-1 RNA motif conserves four bulged-G modules[3][4] (also called E-loops) Bulged-G modules are often associated with intermolecular interactions, and multiple examples are found in ribosomal RNAs. However the biological role that the four bulged-G modules play in the context of Dictyoglomi-1 RNAs remains unknown.
References
- ↑ 1.0 1.1 "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol 11 (3): R31. March 2010. doi:10.1186/gb-2010-11-3-r31. PMID 20230605.
- ↑ "Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis". Nature 462 (7273): 656–659. December 2009. doi:10.1038/nature08586. PMID 19956260. Bibcode: 2009Natur.462..656W.
- ↑ Westhof E (2010). "The amazing world of bacterial structured RNAs". Genome Biol 11 (3): 108. doi:10.1186/gb-2010-11-3-108. PMID 20236470.
- ↑ Lee, JC (2003). Structural studies of ribosomal RNA based on cross-analysis of comparative models and three-dimensional crystal structures (PhD thesis). University of Texas.
External links
 |

