Biology:TMEM241
Transmembrane protein 241 (aka C18orf45, hVVT) is a ubiquitous sugar transporter protein which in humans is encoded by the TMEM241 gene.[1]
Gene
In humans, TMEM241 is a 142,188 bp gene located at 18q11.2 which contains 24 exons.[2]
Gene Neighborhood
TMEM241 is located near CABLES1, RIOK3, and NPC1 on chromosome 18.[3]
mRNA
The primary mRNA for human TMEM241, isoform 1,[3] contains a 5' UTR hairpin loop conserved in primates. The primary mRNA for human TMEM241 isoform 1 contains binding sites in its 3' UTR for the miRNAs 520f-5p, 378a-5p, and 6866-5p.[4]
Protein
General Properties
There are over 10 transcript variants predicted for the human TMEM241 gene found on BLAST. TMEM241 Isoform 1 is approximately 31 kDa in mass. The protein has an isoelectric point of 8.7. and is particularly rich in the amino acid phenylalanine, containing twice the normal proportion of this amino acid.[5]
Conserved Domains
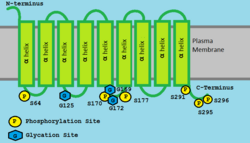
TMEM241 is composed of 9 transmembrane domains forming a hydrophobic integral component of the membrane[2] composed primarily of alpha helices.[6][7][8] TMEM241 contains a VRG4 (Vanadate Resistant Glycosylation[9]) domain with homology to the sugar transporter domain VRG4 from Saccharomyces cerevisiae (yeast).[10]
Post-Translational Modification
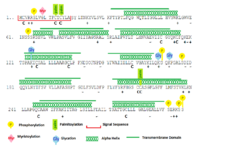
TMEM241 is predicted to undergo various phosphorylations,[11] glycation,[11] palmitoylation.[12] For example, TMEM241 isoform 1[13] has a phosphorylation sites on S6, 64, 170, 177, 291, 295 and 296;[11] glycation sites on K125, 169 and 172;[11] palmitoylation sites on C13, 15, 221.[12]
Interacting Proteins
There is some evidence that this protein may interact with keratin filament based on a two hybrid screen with the keratin protein KRT40.[14]
Expression
TMEM241 is likely to be expressed in all tissues at varying levels from basal to moderate expression.[15] Some studies have found changes in the expression of TMEM241. For instance, in cases of acute megakaryoblastic leukemia, TMEM241 was found to be one of the most upregulated genes.[16] In another case TMEM241 was found to be upregulated during the unfolded protein response following the overexpression of Ero1α (Endoplasmic Reticulum oxidoreduclin 1α).[10]
Homology
TMEM241 is conserved throughout eukaryotes.
Orthologs
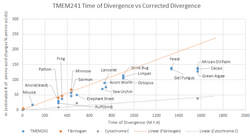
TMEM241 is conserved across all animals and homologs are found throughout eukaryotes. TMEM241 has 19% global identity and 60% identity to GDP-mannose transporter from S. cerevisiae, which contains the VRG4 domain. It is likely that TMEM241 is a GDP-mannose transporter due to this similarity. The graph on the right shows the relative level of conservation of TMEM241 across many species of organism using the principle of a Molecular Clock.
| Scientific name | Common name | Divergence from H. sapiens (m.y.a)[17] | Protein Length (amino acids) | % Identity to H. sapiens[18][19] | Accession number |
|---|---|---|---|---|---|
| Homo sapiens | Human | 0 | 296 | 100 | NP_116322 |
| Mus musculus | Mouse | 90.1 | 297 | 84 | NP_001276595 |
| Calidris pugnax | Ruff (bird) | 320.5 | 296 | 80 | XP_003219697 |
| Python bivittatus | Python | 320.5 | 296 | 74 | XP_014793906 |
| Xenopus laevis | Frog | 354.4 | 293 | 69 | NP_001091222 |
| Salmo salar | Salmon | 436.8 | 296 | 63 | XP_014036453 |
| Octopus bimaculoides | Octopus | 903 | 291 | 35 | XP_014776992 |
| Halyomorpha halys | Stink bug | 903 | 257 | 32 | XP_014284006 |
| Saccharomyces cerevisiae | Yeast | 1335.5 | 216 | 25 | AJS02580 |
| Coccomyxa subellipsoidea | Green Algae | 1570.5 | 296 | 28 | XP_005651133 |
Paralogs
TMEM241 has two paralogs in humans which have homologs throughout eukaryotes, UGTREL8[20] and UGTREL7.[21] TMEM241, UGTREL8 and UGTREL7 are a family of sugar transport proteins with close identity to the GDP-mannose transporter identified in S. cerevisiae.
| % Identity[18][19] | TMEM241
(H.sapiens) |
UGTREL7
(H.sapiens) |
UGTREL8
(H.sapiens) |
GDP Mannose Transporter
(S. cerevisiae) |
|---|---|---|---|---|
| TMEM241 (H. sapiens) | 100% | 19.9% | 23.9% | 19.0% |
| UGTREL7 (H. sapiens) | 19.9% | 100% | 52.2% | 18.8% |
| UGTREL8 (H. sapiens) | 23.9% | 52.2% | 100% | 19.9% |
| GDP Mannose Transporter
(S. cerivisiae) |
19.0% | 18.8% | 19.9% | 100% |
References
- ↑ "Entrez Gene: Transmembrane protein 241". https://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=retrieve&list_uids=85019.
- ↑ 2.0 2.1 "TMEM241 transmembrane protein 241 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene?LinkName=protein_gene&from_uid=109150412.
- ↑ 3.0 3.1 "NCBI NM_032933.5". https://www.ncbi.nlm.nih.gov/nuccore/NM_032933.5.
- ↑ "miRDB - MicroRNA Target Prediction And Functional Study Database". http://mirdb.org/cgi-bin/search.cgi.
- ↑ Brendel, V; Bucher, P; Nourbakhsh, I.R; Blaisdell, B.E; Karlin, S (1992). "Methods and algorithms for statistical analysis of protein sequences". Proc. Natl. Acad. Sci. 89 (6): 2002–2006. doi:10.1073/pnas.89.6.2002. PMID 1549558. Bibcode: 1992PNAS...89.2002B.
- ↑ "SOSUI: Result". http://harrier.nagahama-i-bio.ac.jp/sosui/cgi-bin/adv_sosui.cgi.
- ↑ "Phobius". http://phobius.sbc.su.se/.
- ↑ "DAS-TMfilter server". http://mendel.imp.ac.at/sat/DAS/.
- ↑ "VRG4 | SGD". https://www.yeastgenome.org/locus/S000003193.
- ↑ 10.0 10.1 Hansen, H.G.; Schmidt, J. D.; Soltoft, C. L.; Ramming, T.; Geertz-Hansen, H. M.; Christensen, B.; Sorensen, E. S.; Juncker, A. S. et al. (1 October 2012). "Hyperactivity of the Ero1 Oxidase Elicits Endoplasmic Reticulum Stress but No Broad Antioxidant Response". Journal of Biological Chemistry 287 (47): 39513–39523. doi:10.1074/jbc.M112.405050. PMID 23027870.
- ↑ 11.0 11.1 11.2 11.3 "Welcome to CBS". http://www.cbs.dtu.dk.
- ↑ 12.0 12.1 "CSS-Palm - Palmitoylation Site Prediction". http://csspalm.biocuckoo.org/showResult.php.
- ↑ "transmembrane protein 241 isoform 1 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/109150412.
- ↑ "mentha: the interactome browser". http://mentha.uniroma2.it/.
- ↑ "GDS3834 / 12797 / TMEM241". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS3834:12797.
- ↑ Pelleri, Maria; Piovesan, Allison; Caracausi, Maria; Berardi, Anna; Vitale, Lorenza; Strippoli, Pierluigi (2014). "Integrated differential transcriptome maps of Acute Megakaryoblastic Leukemia (AMKL) in children with or without Down Syndrome (DS)". BMC Medical Genomics 7 (1): 63. doi:10.1186/s12920-014-0063-z. PMID 25476127.
- ↑ Hedges, SB; Marin, J; Suleski, M; Kumar, S. "TimeTree :: The Timescale of Life". http://www.timetree.org.
- ↑ 18.0 18.1 Pearson, W.R. (1990). [5] Rapid and sensitive sequence comparison with FASTP and FASTA. Methods in Enzymology. 183. pp. 63–98. doi:10.1016/0076-6879(90)83007-v. ISBN 9780121820848.
- ↑ 19.0 19.1 "ALIGN". http://seqtool.sdsc.edu/.
- ↑ "UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/NP_055954.1.
- ↑ "SLC35D1 solute carrier family 35 member D1 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/23169.
Further reading
 |