Biology:Proteo-phage-1 RNA motif
From HandWiki
Revision as of 23:43, 1 August 2022 by imported>Gametune (url)
| Proteo-phage-1 | |
|---|---|
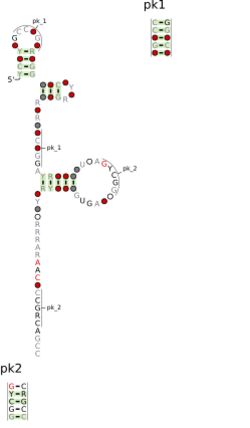 Consensus secondary structure and sequence conservation of Proteo-phage-1 RNA | |
| Identifiers | |
| Symbol | Proteo-phage-1 |
| Rfam | RF03044 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0001263 |
| PDB structures | PDBe |
The Proteo-phage-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] Energetically stable tetraloops often occur in this motif.
Proteo-phage-1 motif RNAs are found in Pseudomonadota. Nearby to most Proteo-phage-1 motif RNAs are genes that are typical of phages. This, in turn, suggests that Proteo-phage-1 motif RNAs are found in prophages, and presumably also used by phage particles, although Proteo-phage-1 motif RNAs have not been observed in genome sequences of purified phages. It is ambiguous whether Proteo-phage-1 RNAs function as cis-regulatory elements or whether they operate in trans.
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
 |

