Biology:Retroposon
Retroposons are repetitive DNA fragments which are inserted into chromosomes after they had been reverse transcribed from any RNA molecule.
Difference between retroposons and retrotransposons
In contrast to retrotransposons, retroposons never encode reverse transcriptase (RT) (but see below). Therefore, they are non-autonomous elements with regard to transposition activity (as opposed to transposons). Non-long terminal repeat (LTR) retrotransposons such as the human LINE1 elements are sometimes falsely referred to as retroposons. However, this depends on the author. For example, Howard Temin published the following definition: Retroposons encode RT but are devoid of long terminal repeats (LTRs), for example long interspersed elements (LINEs). Retrotransposons also feature LTRs and retroviruses, in addition, are packaged as viral particles (virions). Retrosequences are non-autonomous elements devoid of RT. They are retroposed with the aid of the machinery of autonomous elements, such as LINEs; examples are short interspersed nuclear elements (SINEs) or mRNA-derived retro(pseudo)genes.[2][3][4]
Gene duplications
Retroposition accounts for approximately 10,000 gene-duplication events in the human genome, of which approximately 2-10% are likely to be functional.[5] Such genes are called retrogenes and represent a certain type of retroposon.
Horizontal gene transfer
A classical event is the retroposition of a spliced pre-mRNA molecule of the c-Src gene into the proviral ancestor of the Rous sarcoma virus (RSV). The retroposed c-src pre-mRNA still contained a single intron and within RSV is now referred to as v-Src gene.[6]
References
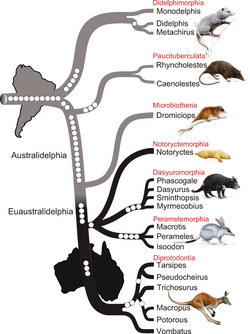
- ↑ Nilsson, M. A.; Churakov, G.; Sommer, M. et al. (2010). "Tracking Marsupial Evolution Using Archaic Genomic Retroposon Insertions". PLOS Biology 8 (7): e1000436. doi:10.1371/journal.pbio.1000436. PMID 20668664.
- ↑ Temin HM (1989). "Retrons in bacteria". Nature 339 (6222): 254–5. doi:10.1038/339254a0. PMID 2471077. Bibcode: 1989Natur.339..254T.
- ↑ "Modulation of Host Genes by Mammalian Transposable Elements". Gene and Protein Evolution. Genome Dynamics. 3 (3rd ed.). Basel: S. Karger. 2007. p. 166. doi:10.1159/000107610. ISBN 978-3-8055-8341-1. OCLC 729848415.
- ↑ "Transposable Elements: Classification, Identification, and Their Use as a Tool for Comparative Genomics". Evolutionary Genomics. Methods in Molecular Biology. 1910. Clifton, NJ. 2019. pp. 185–86. doi:10.1007/978-1-4939-9074-0_6. ISBN 978-3-8055-8341-1. OCLC 145014779.
- ↑ "Extensive gene traffic on the mammalian X chromosome". Science 303 (5657): 537–40. January 2004. doi:10.1126/science.1090042. PMID 14739461. Bibcode: 2004Sci...303..537E.
- ↑ Swanstrom, R; Parker, R C; Varmus, H E; Bishop, J M (May 1983). "Transduction of a cellular oncogene: the genesis of Rous sarcoma virus.". Proceedings of the National Academy of Sciences 80 (9): 2519–2523. doi:10.1073/pnas.80.9.2519. PMID 6302692.
 |