Biology:ATP-dependent Clp protease adaptor protein ClpS
| ClpS | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 clpns with fragments | |||||||||
| Identifiers | |||||||||
| Symbol | ClpS | ||||||||
| Pfam | PF02617 | ||||||||
| InterPro | IPR003769 | ||||||||
| SCOP2 | 1mbx / SCOPe / SUPFAM | ||||||||
| |||||||||
ClpS is an N-recognin in the N-end rule pathway.[1] ClpS interacts with protein substrates that have a bulky hydrophobic residue (leucine, phenylalanine, tyrosine, and tryptophan) at the N-terminus. The protein substrate is then degraded by the ClpAP protease.[2][3]
In molecular biology, the ATP-dependent Clp protease adaptor protein ClpS is a bacterial protein. In the bacterial cytosol, ATP-dependent protein degradation is performed by several different chaperone-protease pairs, including ClpAP. ClpS directly influences the ClpAP machine by binding to the N-terminal domain of the chaperone ClpA. The degradation of ClpAP substrates, both SsrA-tagged proteins and ClpA itself, is specifically inhibited by ClpS. ClpS modifies ClpA substrate specificity, potentially redirecting degradation by ClpAP toward aggregated proteins.[4]
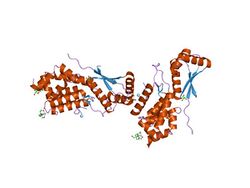
ClpS is a small alpha/beta protein that consists of three alpha-helices connected to three antiparallel beta-strands.[5] The protein has a globular shape, with a curved layer of three antiparallel alpha-helices over a twisted antiparallel beta-sheet. Dimerization of ClpS may occur through its N-terminal domain. This short extended N-terminal region in ClpS is followed by the central seven-residue beta-strand, which is flanked by two other beta-strands in a small beta-sheet.
See also
References
- ↑ "The N-end rule pathway and regulation by proteolysis". Protein Science 20 (8): 1298–345. August 2011. doi:10.1002/pro.666. PMID 21633985.
- ↑ "The N-end rule pathway". Annual Review of Biochemistry 81: 261–89. 10 April 2012. doi:10.1146/annurev-biochem-051710-093308. PMID 22524314.
- ↑ "ClpS is an essential component of the N-end rule pathway in Escherichia coli". Nature 439 (7077): 753–6. February 2006. doi:10.1038/nature04412. PMID 16467841. Bibcode: 2006Natur.439..753E.
- ↑ "ClpS, a substrate modulator of the ClpAP machine". Molecular Cell 9 (3): 673–83. March 2002. doi:10.1016/S1097-2765(02)00485-9. PMID 11931773.
- ↑ "Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA". Nature Structural Biology 9 (12): 906–11. December 2002. doi:10.1038/nsb869. PMID 12426582.
 |

