Biology:Aichivirus A
| Aichivirus A | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Picornavirales |
| Family: | Picornaviridae |
| Genus: | Kobuvirus |
| Species: | Aichivirus A
|
Aichivirus A formerly Aichi virus (AiV)[1] belongs to the genus Kobuvirus in the family Picornaviridae.[2] Six species are apart of the genus Kobuvirus, Aichivirus A-F.[3] Within Aichivirus A, there are six different types including human Aichi virus, canine kobuvirus, murine kobuvirus, Kathmandu sewage kobuvirus, roller kobuvirus, and feline kobuvirus.[3] Three different genotypes are found in human Aichi virus, represented as genotype A, B, and C.[3]
AiV is a non-enveloped positive sense ssRNA virus with icosahedral morphology.[3] Aichivirus A was originally identified after a 1989 outbreak of acute gastroenteritis in the Aichi Prefecture that was linked to raw oyster consumption per genetic analysis.[1][4][5] Human Aichi Virus can cause gastroenteritis with symptoms arising such as vomiting, diarrhea, abdominal pain, nausea, and feve.[3][6]
Aichivirus A can be found in a variety of environmental areas including sewage, groundwater, river water, and shellfish.[2] Aichivirus A is present in many world regions, and in sometimes greater abundance than other well-known enteric viruses.[2] Aichiviruses have been seen in Asia, Europe, South America, and Africa.[2] It has since been isolated in populations of Finland children,[7] Pakistani children, and Japan ese travelers.[8] The widespread nature of aichivirus A can be seen in the high percentage of AiV antibodies in adult human populations found in several countries.[3]
Transmission occurs through the fecal-oral route.[2] After the virus is replicated in the gastrointestinal tract, the pathogen can be found in fecal samples of infected individuals.[2] Water and shellfish contaminated with human sewage can propagate aichivirus A.[2]
Discovery
Aichivirus A was first characterized after an outbreak of gastroenteritis in the Aichi Prefecture of Japan, this region is where the name of the virus was derived from.[5] Fecal samples from infected individuals were taken and transported to a lab where they described the novel virus.[5] These viral particles were 30 nm in diameter, a spherical shape, and cytopathic for BSC-1 cells (kidney cells of African green monkey).[3][5] The infection was attributed to contaminated raw oyster found in vinegar.[5]
Aichivirus A has been seen and described across many Asian countries, however the first appearance of aichivirus outside of this region was isolated in Europe and South America in 2006.[9] Through genetic analysis of isolates from Brazil and Germany, the nucleotide sequences were found to be similar to known Aichivirus nucleotide sequences.[9] Notably, the German strain appeared to be of genotype A and the Brazil strain appeared to be of genotype B.[9] Screening in Germany for antibodies to Aichivirus displayed a seroprevalence of 76%, which is comparable to seroprevalence in Japan.[9] Therefore, European infection with Aichivirus is as common as it is in Asia.[9]
Human infection
Propagation
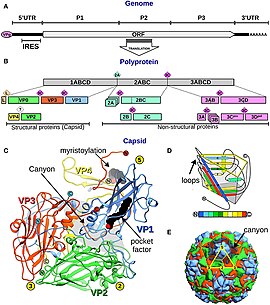
Aichivirus A enters host cells through receptor-mediated endocytosis, a cellular uptake mechanism.[3] After viral attachment and entry, the virion particle is uncoated releasing the genome into the cytoplasm.[3] Similar to other viruses within the Picornaviridae family, viral replication and translation occurs in the cytoplasm.[10] The positive sense ssRNA is directly translated into protein by the host cell ribosomes, while some of the ssRNA is used as a template to replicate the viral genome. Capsid proteins, L protein, nonstructural proteins, and stable intermediates are produced after the polyprotein is processed. Protein production is directly related to synthesis of plus-strand RNA replication complex.[10][3] The plus-strand RNA genome is packaged into the assembled viral particle, along with VpG (Viral genomic protein).[3] A completed viral particle has 60 capsid proteins copies made up of 12 pentamers.[3] The pentamer is made up by the 5S subunit composed of VP0, VP1, and VP3 protein aggregates.[3] After the viral particle is assembled, it is released from host cells by cell lysis, making Aichivirus A a lytic virus.[3][11]
Characteristics
Most aichivirus A infection in humans are mild, asymptomatic infections lasting between 48–72 hours.[3] However, it can develop into the common symptoms of gastroenteritis: fever, nausea, vomiting, abdominal pain.[12][3] Viral replication in the gastrointestinal tract damages the enterocyte layer in the intestinal villi interfering with water reabsorption[12] This can lead to the symptoms appearing with infection.
Aichivirus A can become an opportunistic pathogen in those with HIV and is seen in high levels in the feces of those with HIV.[13][14] Aichivirus A is also suspected as an opportunistic pathogan in those with X-linked agammaglobulinemia.[14] Aichivirus is an emerging pathogen in those with B-cell deficiencies, however there is no explanation why[14] In patients with primary immune deficiencies, chronic aichivirus infection can cause immunodysregulation.[15] Human aichivirus was deemed to effect multiple organs leading to the clinical symptom presentation.[15] The aichivirus genome was detected in symptomatic patients and in infected organs, while was not seen in asymptomatic individuals.[15] Notably, in Japan there is a correlation with aichivirus A infection and lower respiratory tract disease.[16]
Genome
The RNA genome of AiV A is composed of 8280 nucleotides. Along the 5' end of the RNA, there is an untranslated region consisting of 744 nucleotides, a VpG protein, and an internal ribosomal entry site (IRES). Following the 5' untranslated region, the open reading frame is approximately 7.3kB consisting of 2432 amino acids. The L protein, leader peptide, is the first protein translated within the polypeptide, followed by the structural proteins, and then nonstructural proteins. Cleavage into the different proteins occurs by viral proteases.[17] The capsid proteins are made up of three segments in the RNA: VP0, VP3, and VP1.[3][17] These capsid proteins together are known as the P1 region on the genome.[3] The encoded capsid proteins form a protomer that form into 12 pentamers during self-assembly.[17][18] X-ray crystallography of human aichi virus virion structure determined that the VP3 knob structure and VP0 surface loop are smaller compared to other viruses in picornaviruses.[17] P2 and P3 are the regions of the RNA genome that are the non-structural proteins involved in replication control.[17][19] For example, the protein 3D within the P3 region encodes for the viral RNA-dependent RNA polymerase used in replication.[20] The 3B protein also in the P3 region encodes for the VpG protein, which is important promoting replication. Following the P2 and P3 region, there is a 3' untranslated region of about 237 nucleotides and a poly-A tail.[3]
Genotypic differences
Differences at the 3CD nonstructural protein junction in the viral genome results in distinct genotypic differences.[21][3] When comparing the junction between the C-terminus of the 3C region and the N-terminus of the 3D region, three distinct genotypic types are seen.[3] In studies, there appeared to be a geographical distribution to the genotypes. In some countries genotype B is prevalent, while in others genotype A dominates. In Finland and Spain genotype A was more prevalent.[7][22] However, in China, Bangladesh, and Pakistan genotype B is more widely seen in gastroenteritis outbreaks.[23][24] However, genotype C is not widely seen to cause human infection and has only been described in one study a fecal sample from a child case of gastroenteritis after a trip to Mali.[3][21] The VP1 region is used to classify the picornaviruses and can also be used to differentiate between aichivirus A genotypes.[3] Some studies have seen more variation in th VP1 region and suggest that this region may be a better region to differentiate between genotypes.[9]
Environmental occurrence
Aichivirus A can be found in a variety of environmental sources potentially leading to infection through food and water consumption. Aichivirus A causes infection through the fecal-oral route, where contaminated food and water sources are ingested.[3] Some studies suggest using Aichivirus A as a method to detect viral contamination in environmental samples.[2]
Shellfish
Enteric viruses can propagate through bivalve mollusks which filter surrounding water for food and retain enteric viruses.[3] Many safety protocols only take into account bacteria and not viruses, which makes shellfish a vector for viral transmission.[25] Aichivirus A was first detected in an outbreak due to contaminated oysters, and contaminated seafood has been associated with aichivrius A outbreaks worldwide.[3] In a year-long study in Japan on viral detection in clams, 33% of the grocery store samples contained aichivirus A.[26]
Sewage
Aichivirus A has been reported at high rates in wastewater but was first seen in 2010.[27] Wastewater treatment cannot get rid of all the viral particles before being discharged into the environment.[3] Due to the stability of aichivirus in sewage before and after treatment, aichivirus A is likely a human fecal pollutant indicator.[3] Aichivirus A has been detected in wastewater in America, Europe, Africa, and Asia.[3] In one study, samples of treated sewage contained a 91.7% prevalence of aichivirus A.[28]
River and ground water
River water and ground water can be a reservoir for aichivirus A, due to viruses not being removed during the natural filtration cycle.[3] Aichivirus A was first studied in river water in Venezuela in 2010 with detection in 45% of samples.[27] Aichivirus A has since been detected in water sources worldwide, including in tap water and ground water in America.[3]
Research
Under an electron microscope, Aichivirus A appears as a small, round virus making it hard to distinguish it from other viruses with a similar morphology.[16] Under electron microscopy, a canyon-like valley is seen on the surface of the capsid, likely where receptor binding occurs for entry.[3] The viral particle is stable in acidic conditions until a pH of 2 and remains stable under known experimental methods to disrupt the viral particle.[29] These methods include heat, hydrostatic pressure, and detergent conditions.[29][3] In human cell lines like HeLa, a cytopathic effect is not seen, however a cytopathic effect is seen in BSC-1 cell lines and Vero Cells.[3]
An enzyme-linked immunoabsorbant assay (ELISA) has been developed to detect aichivirus A antigens.[16] Reverse transcription-RNA polymerase chain reaction is also widely used in aichivirus research for identification and genotype differentiation.[30] A loop-mediated isothermal amplification (LAMP) assay has been created for aichivirus A to be used in water samples.[31] The LAMP assay allows for a rapid and specific detection of aichivirus A.[31] Reverse transcription-quantitative PCR (RT-qPCR) is also widely used for detection and to determine viral numbers.[3]
References
- ↑ 1.0 1.1 "Phylogeny and prevalence of kobuviruses in dogs and cats in the UK". Veterinary Microbiology 164 (3–4): 246–252. June 2013. doi:10.1016/j.vetmic.2013.02.014. PMID 23490561.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 "Aichi virus 1: environmental occurrence and behavior". Pathogens 4 (2): 256–268. May 2015. doi:10.3390/pathogens4020256. PMID 25996404.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 3.24 3.25 3.26 3.27 3.28 3.29 3.30 3.31 3.32 3.33 3.34 "A Comprehensive Review on Human Aichi Virus". Virologica Sinica 35 (5): 501–516. October 2020. doi:10.1007/s12250-020-00222-5. PMID 32342286.
- ↑ "Aichi virus (AiV)". A Dictionary of Virology (3rd ed.). San Diego, California: Academic Press. 2001. pp. 9. ISBN 978-0-12-465327-6. https://books.google.com/books?id=vYotjPWL_2IC&pg=PA9.
- ↑ 5.0 5.1 5.2 5.3 5.4 Viral Gastroenteritis. Gulf Professional Publishing. 2003. pp. 645–. ISBN 978-0-444-51444-8. https://books.google.com/books?id=crOf-W3g-MQC&pg=PA645.
- ↑ "Identification of Aichi virus infection by measurement of immunoglobulin responses in an enzyme-linked immunosorbent assay". Journal of Clinical Microbiology 39 (11): 4178–4180. November 2001. doi:10.1128/JCM.39.11.4178-4180.2001. PMID 11682554.
- ↑ 7.0 7.1 "Aichi virus infection in children with acute gastroenteritis in Finland". Epidemiology and Infection 138 (8): 1166–1171. August 2010. doi:10.1017/S0950268809991300. PMID 19961643.; Lay summary in: "Aichi virus infection in children with acute gastroenteritis in Finland". Issues in Global, Public, Community, and Institutional Health. ScholarlyEditions. 2011. p. 793. ISBN 978-1-4649-6382-7. https://books.google.com/books?id=Txe-WGjCm-oC&pg=PT793.
- ↑ "Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia". Microbiology and Immunology 39 (6): 433–435. 1995. doi:10.1111/j.1348-0421.1995.tb02225.x. PMID 8551977.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 "Molecular characterization of the first Aichi viruses isolated in Europe and in South America". Archives of Virology 151 (6): 1199–1206. June 2006. doi:10.1007/s00705-005-0706-7. PMID 16421634.
- ↑ 10.0 10.1 "Picornavirus Entry" (in en). Viral Entry into Host Cells. Advances in Experimental Medicine and Biology. 790. New York, NY: Springer. 2013. pp. 24–41. doi:10.1007/978-1-4614-7651-1_2. ISBN 978-1-4614-7651-1.
- ↑ "Picornaviridae-the ever-growing virus family". Archives of Virology 163 (2): 299–317. February 2018. doi:10.1007/s00705-017-3614-8. PMID 29058149.
- ↑ 12.0 12.1 "Enterically infecting viruses: pathogenicity, transmission and significance for food and waterborne infection". Journal of Applied Microbiology 98 (6): 1354–1380. June 2005. doi:10.1111/j.1365-2672.2005.02635.x. PMID 15916649.
- ↑ "Unexplained diarrhoea in HIV-1 infected individuals". BMC Infectious Diseases 14: 22. January 2014. doi:10.1186/1471-2334-14-22. PMID 24410947.
- ↑ 14.0 14.1 14.2 "Aichivirus: an Emerging Pathogen in Patients with Primary and Secondary B-Cell Deficiency". Journal of Clinical Immunology 43 (3): 532–535. April 2023. doi:10.1007/s10875-022-01410-6. PMID 36449139.
- ↑ 15.0 15.1 15.2 "Chronic Aichi Virus Infection As a Cause of Long-Lasting Multiorgan Involvement in Patients With Primary Immune Deficiencies". Clinical Infectious Diseases 77 (4): 620–628. August 2023. doi:10.1093/cid/ciad237. PMID 37078608.
- ↑ 16.0 16.1 16.2 "Prevalence of newly isolated, cytopathic small round virus (Aichi strain) in Japan". Journal of Clinical Microbiology 31 (11): 2938–2943. November 1993. doi:10.1128/jcm.31.11.2938-2943.1993. PMID 8263178.
- ↑ 17.0 17.1 17.2 17.3 17.4 "Structure of Aichi Virus 1 and Its Empty Particle: Clues to Kobuvirus Genome Release Mechanism". Journal of Virology 90 (23): 10800–10810. December 2016. doi:10.1128/JVI.01601-16. PMID 27681122.
- ↑ "In vitro synthesis and assembly of picornaviral capsid intermediate structures". Journal of Virology 44 (3): 900–906. December 1982. doi:10.1128/jvi.44.3.900-906.1982. PMID 6294338.
- ↑ "Structure of human Aichi virus and implications for receptor binding". Nature Microbiology 1 (11): 16150. September 2016. doi:10.1038/nmicrobiol.2016.150. PMID 27595320. https://ora.ox.ac.uk/objects/uuid:cff9ca7d-90b1-48de-a794-db2258ec066a.
- ↑ "VI, 3. Molecular biology and epidemiology of Aichi virus and other diarrhoeogenic enteroviruses". Perspectives in Medical Virology 9: 645–657. 2003. doi:10.1016/S0168-7069(03)09040-2. ISBN 9780444514448. PMID 32336843.
- ↑ 21.0 21.1 "Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients". Journal of Clinical Microbiology 46 (4): 1252–1258. April 2008. doi:10.1128/JCM.02140-07. PMID 18256215.
- ↑ "Epidemiology of Aichi virus in fecal samples from outpatients with acute gastroenteritis in Northwestern Spain". Journal of Clinical Virology 118: 14–19. September 2019. doi:10.1016/j.jcv.2019.07.011. PMID 31382225.
- ↑ "Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam". Journal of Clinical Microbiology 45 (7): 2287–2288. July 2007. doi:10.1128/JCM.00525-07. PMID 17522267.
- ↑ "Aichi virus strains in children with gastroenteritis, China". Emerging Infectious Diseases 15 (10): 1703–1705. October 2009. doi:10.3201/eid1510.090522. PMID 19861087.
- ↑ "Detection and quantification of hepatitis A virus and norovirus in Spanish authorized shellfish harvesting areas". International Journal of Food Microbiology 193: 43–50. January 2015. doi:10.1016/j.ijfoodmicro.2014.10.007. PMID 25462922.
- ↑ "Detection of human enteric viruses in Japanese clams". Journal of Food Protection 71 (8): 1689–1695. August 2008. doi:10.4315/0362-028X-71.8.1689. PMID 18724766.
- ↑ 27.0 27.1 "Molecular detection and characterization of Aichi viruses in sewage-polluted waters of Venezuela". Applied and Environmental Microbiology 76 (12): 4113–4115. June 2010. doi:10.1128/AEM.00501-10. PMID 20418428. Bibcode: 2010ApEnM..76.4113A.
- ↑ "Prevalence and genetic diversity of Aichi viruses in wastewater and river water in Japan". Applied and Environmental Microbiology 77 (6): 2184–2187. March 2011. doi:10.1128/AEM.02328-10. PMID 21257803. Bibcode: 2011ApEnM..77.2184K.
- ↑ 29.0 29.1 "Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans". Journal of Virology 72 (10): 8408–8412. October 1998. doi:10.1128/JVI.72.10.8408-8412.1998. PMID 9733894.
- ↑ "Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the Picornavirus family associated with gastroenteritis in humans". Journal of Clinical Microbiology 38 (8): 2955–2961. August 2000. doi:10.1128/JCM.38.8.2955-2961.2000. PMID 10921958.
- ↑ 31.0 31.1 "Development of Rapid and Specific Detection for the Human Aichivirus A Using the Loop-Mediated Isothermal Amplification from Water Samples". Indian Journal of Microbiology 59 (3): 375–378. September 2019. doi:10.1007/s12088-019-00803-3. PMID 31388217.
Wikidata ☰ Q18205171 entry
 |