Biology:Picornavirales
From HandWiki
Short description: Order of viruses
| Picornavirales | |
|---|---|
| |
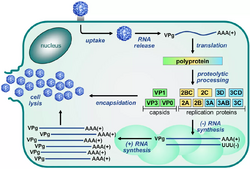
| Picornavirus replication cycle | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Picornavirales |
Picornavirales is an order of viruses with vertebrate, invertebrate, protist and plant hosts.[1] The name has a dual etymology.[2] First, picorna- is an acronym for poliovirus, insensitivity to ether, coxsackievirus, orphan virus, rhinovirus, and ribonucleic acid.[2] Secondly, pico-, meaning extremely small, combines with RNA to describe these very small RNA viruses.[2] The order comprises viruses that historically are referred to as picorna-like viruses.
Characteristics
The families within this order share a number of common features:
- The virions are non-enveloped, icosahedral, and about 30 nanometers in diameter.
- The capsid has a "pseudo T=3" structure, and is composed of 60 protomers each made of three similar-sized but nonidentical beta barrels.
- The genome is made of one or a few single-stranded RNA(s) serving directly as mRNA, without overlapping open reading frames.
- The genome has a small protein, VPg, covalently attached to its 5' end, and usually a poly-adenylated 3' end.
- Each genome RNA is translated into polyprotein(s) yielding mature viral proteins through one or several virus-encoded proteinase(s).
- A hallmark of the Picornavirales is a conserved module of sequence domains, Hel-Pro-Pol, which is typical of (from the amino- to the carboxy-end of the polyprotein):
- A helicase belonging to superfamily III
- [the VPg is encoded between these two domains]
- A chymotrypsin-like protease where the catalytic residue is typically a cysteine rather than a serine,
- A polymerase belonging to superfamily I; this conserved module is a hallmark of the Picornavirales
The evolution of picorna-like viruses seems to have antedated the separation of eukaryotes into the extant crown groups.[3]
Taxonomy
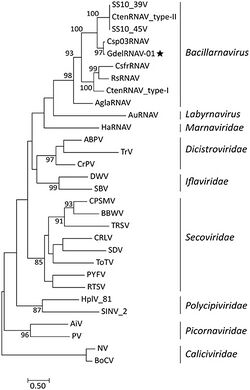
The following families are recognized:[4]
- Caliciviridae
- Dicistroviridae
- Iflaviridae
- Marnaviridae
- Picornaviridae
- Polycipiviridae
- Secoviridae
- Solinviviridae
References
- ↑ Le Gall, Olivier; Christian, Peter; Fauquet, Claude M.; King, Andrew M. Q.; Knowles, Nick J.; Nakashima, Nobuhiko; Stanway, Glyn; Gorbalenya, Alexander E. (2008-04-01). "Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture". Archives of Virology 153 (4): 715–27. doi:10.1007/s00705-008-0041-x. PMID 18293057.
- ↑ Jump up to: 2.0 2.1 2.2 "Picornaviridae" (in en). October 2017. https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/picornavirales/w/picornaviridae.
- ↑ Koonin, Eugene V.; Wolf, Yuri I.; Nagasaki, Keizo; Dolja, Valerian V. (2008). "The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups". Nature Reviews Microbiology 6 (12): 925–939. doi:10.1038/nrmicro2030. PMID 18997823.
- ↑ "Virus Taxonomy: 2019 Release". International Committee on Taxonomy of Viruses. https://ictv.global/taxonomy.
External links
Wikidata ☰ Q6553 entry
 |