Biology:Avian HBV RNA encapsidation signal epsilon
| AHBV_epsilon cis-regulatory RNA element | |
|---|---|
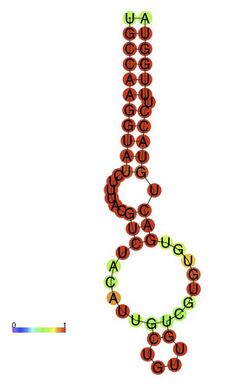 Predicted secondary structure and sequence conservation of AHBV_epsilon | |
| Identifiers | |
| Symbol | AHBV_epsilon |
| Rfam | RF01313 |
| Other data | |
| RNA type | Cis-reg |
| Domain(s) | Viruses |
| GO | 0019079 |
| SO | 0005836 |
| PDB structures | PDBe |
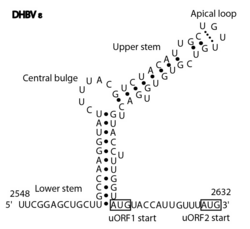
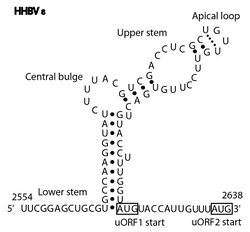
The Avian HBV RNA encapsidation signal epsilon (AHBV epsilon[1]) is an RNA structure that is shown to facilitate encapsidation of the pregenomic RNA required for viral replication. There are two main classes of encapsidation signals in avian hepatitis B viruses - Duck hepatitis B virus (DHBV) and Heron hepatitis B virus (HHBV) like. DHBV is used as a model to understand human Hepatitis B virus. Although studies have shown that the HHBV epsilon has less pairing in the upper stem than DHBV, this pairing is not absolutely required for DHBV infection in ducks.[2]
DHBV epsilon consists of a stem structure, a bulge and an apical loop (Fig. 1). The RNA structure was determined by chemical probing, NMR analysis and by mutagenesis.[3]
HHBV epsilon also consists of a stem structure, a bulge and an apical loop (Fig. 2). The RNA structure has been determined by chemical probing and by mutagenesis. However, it has less base pairing in the upper stem than DHBV epsilon. [4]
Recent NMR analysis of both the HHBV and DHBV epsilon have shown that the apical loop to be capped by a stable well-structured UGUU tetraloop.[5]
In the Rfam database a single covariation model (AHBV_epsilon box right) best discriminates between the true instances of the element in sequence databases and background. The secondary structure diagram is a computer generated view of the structure with highly conserved bases shown in red. It includes the major elements shown in Figures 1 and 2.[citation needed]
See also
- HBV RNA encapsidation signal epsilon
References
- ↑ "Rfam: Family: AHBV_epsilon (RF01313)". http://rfam.org/family/RF01313.
- ↑ "A high level of mutation tolerance in the multifunctional sequence encoding the RNA encapsidation signal of an avian hepatitis B virus and slow evolution rate revealed by in vivo infection". Journal of Virology 85 (18): 9300–9313. September 2011. doi:10.1128/JVI.05005-11. PMID 21752921.
- ↑ "The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function". Nucleic Acids Research 26 (21): 4818–4827. November 1998. doi:10.1093/nar/26.21.4818. PMID 9776740.
- ↑ "Sequence- and structure-specific determinants in the interaction between the RNA encapsidation signal and reverse transcriptase of avian hepatitis B viruses". Journal of Virology 71 (7): 4971–4980. July 1997. doi:10.1128/JVI.71.7.4971-4980.1997. PMID 9188560.
- ↑ "Thermodynamics and NMR studies on Duck, Heron and Human HBV encapsidation signals". Nucleic Acids Research 35 (8): 2800–2811. 2007. doi:10.1093/nar/gkm131. PMID 17430968.
External links
 |