Biology:C8orf48
 Generic protein structure example |
C8orf48 is a protein that in humans is encoded by the C8orf48 gene.[1] C8orf48 is a nuclear protein specifically predicted to be located in the nuclear lamina.[2][3] C8orf48 has been found to interact with proteins that are involved in the regulation of various cellular responses like gene expression, protein secretion, cell proliferation, and inflammatory responses.[4][5][6] This protein has been linked to breast cancer and papillary thyroid carcinoma.[7][8]
Gene
C8orf48 is located on chromosome 8 (8p22) and spans from 13,566,843 to 13,568,288 on the positive strand.[9] C8orf48 has an exon count of 1 and no introns.[1][10] This protein does not have any isoforms nor exhibit any alternative splicing.
Protein
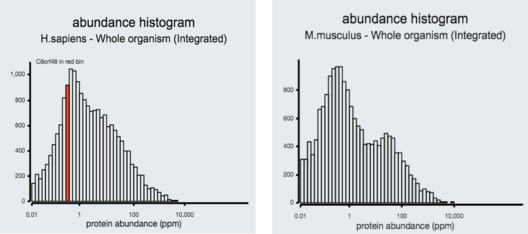
The protein C8orf48 is 319 amino acids in length.[11] The molecular weight of this protein is 36.9 kDa and the isoelectric point is 8.86.[12] The C8orf48 protein is predicted to be a nuclear protein particularly located in the nuclear lamina.[2] This protein does not possess any signal peptides or transmembrane domains. This protein has also been found to be fairly abundant in humans.[13]
Structure
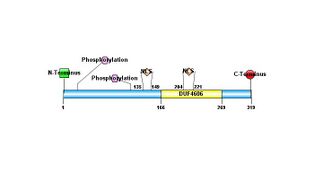
C8orf48 protein has two predicted nuclear localization signals one spanning from 135-149 amino acids and the other from 204-221.[3] The secondary structure of C8orf48 protein is composed of primarily alpha-helices and coiled coils. The structure is composed of very little beta sheets, a total of three areas demonstrate possible beta sheet structure.[12]
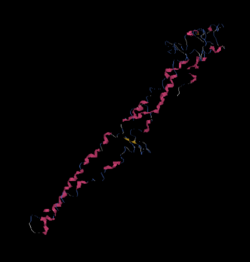
The Tertiary structure of C8orf48 was obtained from the iTASSER program.
Post-translational modifications
C8orf48 has various predicted post-translational modifications. These post-translational modifications include O-glycosylation, Glycation, N-linked glycosylation, Phosphorylation Sites, Yin-Yang sites, sumoylation, and SUMO interactions.[15]
Subcellular localization
PSORT II results determined that the protein C8orf48 does not have a signalPeptide as well as no transmembrane domains.[16] The prediction is that C8orf48 is most likely nuclear and potentially cytoplasmic. When comparing orthologs of C8orf48 we see very similar results, the protein is predicted to be localized in the nucleus predominantly and secondly predicted to be localized in the cytoplasm. Further sub-cellular localization analysis was done through the use of CELLO.[17] These results also support the notion that C8orf48 is localized in the nucleus.
Homology
C8orf48 is conserved in mammals, amphibians, reptiles, aves, and fish.[18] C8orf48 orthologs were unable to be found in bacteria, archea, plants, and fungi.[18] There were no human paralogs found of C8orf48. Certain portions of the DUF 4606 domain is highly conserved in the orthologs.
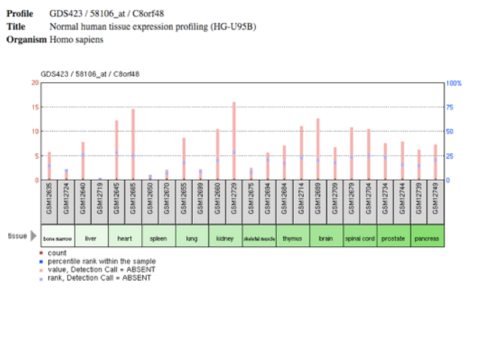
Expression
This gene has been found to be overly expressed in the tissues of the testis and colon muscle, as well as expressed in 76 developmental stages.[19] C8orf48 has been found to be expressed most often in the bladder, bone, heart, larynx, testis, and thyroid.[20] In regards to the developmental stages, C8orf48 was most often found in the embroid body.[20]
GEO profiles
In a study regarding multiple myeloma bone marrow mesenchymal stromal cells shows that the expression of C8orf48 is lower in the disease state cells in comparison to healthy cells.[21] the opposite is demonstrated in the GeoProfile regarding Endometriosis, in this study, it was found that C8orf48 levels are higher in the disease state than in the healthy state.[22] Other studies demonstrate differential expression of Papillary Thyroid cancer and Estrogen Receptor alpha-silenced MCF7 breast cancer cells. In control samples the levels of C8orf48 were lower than that of those with the Estrogen receptor knockdown.[7]
Regulation of expression
The transcription factors that act on C8orf48 are presented in Table 1. The majority of the transcription factors are involved in cell growth, proliferation, or regulation of cell migration. This implies that C8orf48 may play a role in the cell cycle. A few of the transcription factors presented themselves more than once, on both the positive and negative strand. These transcription factors include MAX binding protein and Estrogen-related receptor alpha (secondary DNA binding preference) both of which are involved in cell growth.[23][24]
| Transcription Factor | Function |
| Myeloid zinc finger protein MZF1 | Found to be expressed in hematopoietic progenitor cells that are committed to myeloid lineage differentiation. It contains 13 C2H2 zinc fingers arranged in two domains that are separated by a short glycine- and proline-rich sequence.[25] |
| Zinc finger with KRAB and SCAN domains 3 | It is mainly located in the nucleus although during starvation periods it relocates to the cytoplasm. This allows for expression of target genes involved in autophagy and lysosome biogenesis/function. Acts as a repressor of autophagy and has been found and promote cancer cell progression and/or migration in various tumors and myelomas.[26] |
| T-box transcription factor TBX20 | Found to be involved in the regulation of a genetic program for cranial motor neuron cell body migration.[27] |
| Kruppel-like zinc finger protein 219 | Found to be involved in the regulation of chondrocyte differentiation by assembling a transcription factory with Sox9.[28] |
| KRAB-containing zinc finger protein 300 | |
| Krueppel-like factor 2 (lung) (LKLF) | KLF2 regulates T-cell trafficking by promoting the expression of S1P1 that is a lipid-binding receptor. This transcription factor binds to the CACCC box in the promoter sequence.[29] |
| Estrogen-related receptor alpha (secondary DNA binding preference) | |
| AREB6 (Atp1a1 regulatory element binding factor 6) | AREB6’s structure is composed of two zinc-finger clusters in N- and C-terminal regions, and one homeodomain in the middle. It has been found to regulate the expression of the Na, K-ATPase α1 subunit, interleukin 2 and δ-crystallin genes.[30] |
| MAX binding protein | Transcriptional repressor and an antagonist of Myc-dependent transcriptional activation and cell growth.[23] |
| Leucine rich repeat (in FLII) interacting protein 1 | A transcriptional repressor that potentially regulates TNF, EGFR and PDGFA. Possibly involved in the control of smooth muscle cell proliferation following artery injury.[31] |
| E2F transcription factor 1 | Plays an important role in the control of the cell cycle and tumor suppressor proteins. Also found to be the target of transforming proteins of small DNA tumor viruses. Lastly, it can mediate cell proliferation and p53-dependent/independent apoptosis.[32] |
| Kruppel-like factor 7 (ubiquitous, UKLF) | Members of the family it pertains to are responsible for the regulation of cell proliferation, differentiation, and survival. Found to possibly contribute to the progression of type 2 diabetes by inhibiting the expression of insulin, secretion in pancreatic beta-cells, and by deregulating adipocytokine secretion in adipocytes.[33] |
| CCAAT/enhancer binding protein (C/EBP), epsilon | Important for terminal differentiation and functional maturation of committed granulocyte progenitor cells. Mutations in the gene that encodes this protein has been found to be associated with Specific Granule Deficiency.[34] |
| Zinc finger / POZ domain transcription factor | The POZ domain is a conserved protein-protein interaction motif found in transcription factors, oncogenic proteins, ion channel proteins, and some actin-associated proteins.[35] |
| Oligodendrocyte lineage transcription factor 2 | Tends to be expressed in oligodendrocyte tumors in the brain. This protein is an essential regulator of ventral neuroectodermal progenitor cell fate and chromosomal translocation t(14;21)(q11.2;q22) associated with T-cell acute lymphoblastic leukemia.[36] |
| Basic krueppel-like factor (KLF3) | One of its related pathways is transcriptional regulation in cancer, and may possibly have a role in hematopoiesis. Binds to the CACCC box of erythroid cell-expressed genes.[37] |
Protein interactions
The proteins that interact with C8orf48 include Deleted In Liver Cancer 1 Protein (DLC1), MyoD Family Inhibitor (MDFI), Zinc Finger Protein 14 (ZNF14), and Sacroglycan Zeta (SGCZ).[4] All of these protein interactions were found experimentally via a two-hybrid pooling approach, two-hybrid array, or two-hybrid screen.[6]
Clinical significance
C8orf48 has been found in studies regarding various types of carcinoma. Different C8orf48 expression levels have been found in Papillary Thyroid cancer and Estrogen Receptor alpha-silenced MCF7 breast cancer cells.[21][22]
References
- ↑ 1.0 1.1 "C8orf48 chromosome 8 open reading frame 48 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/157773.
- ↑ 2.0 2.1 "Subnuclear compartments prediction system". http://array.bioe.uic.edu/subnuclear.htm.
- ↑ 3.0 3.1 "Motif Scan". http://myhits.isb-sib.ch/cgi-bin/motif_scan.
- ↑ 4.0 4.1 "STRING: functional protein association networks". http://string-db.org/cgi/network.pl?taskId=VW1u1gSMmsPV.
- ↑ "Protein Kinase C Signaling | Cell Signaling Technology". http://www.cellsignal.com/contents/science-pathway-research-ca-%20camp-and-lipid-signaling/protein-kinase-c-signaling/pathways-kinase-c.
- ↑ 6.0 6.1 PSICQUIC. "PSICQUIC View". http://www.ebi.ac.uk/Tools/webservices/psicquic/view/results.xhtml?conversationContext=2.
- ↑ 7.0 7.1 "GDS4061 / 236634_at". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS4061:236634_at.
- ↑ "GDS1665 / 236634_at". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS1665:236634_at.
- ↑ "Genomatix: ElDorado Result". https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=36d50476195bb39487a091ed603aba43;RESULT=C8orf48.[yes|permanent dead link|dead link}}]
- ↑ "Transcript: C8orf48-001 (ENST00000297324) - Summary - Homo sapiens - Ensembl genome browser 84". http://www.ensembl.org/Homo_sapiens/Transcript/Summary?g=ENSG00000164743;r=8:13566843-13568287;t=ENST00000297324.
- ↑ "Homo sapiens chromosome 8 open reading frame 48 (C8orf48), mRNA - Nucleotide - NCBI". https://www.ncbi.nlm.nih.gov/nuccore/NM_001007090?report=genbank.
- ↑ 12.0 12.1 "Methods and algorithms for statistical analysis of protein sequences". Proceedings of the National Academy of Sciences of the United States of America 89 (6): 2002–6. 1992. doi:10.1073/pnas.89.6.2002. PMID 1549558. Bibcode: 1992PNAS...89.2002B.
- ↑ "PaxDb". http://pax-db.org/#!protein/9606.ENSP00000297324.
- ↑ "I-TASSER server for protein structure and function prediction". http://zhanglab.ccmb.med.umich.edu/I-TASSER/.
- ↑ "CBS Prediction Servers". http://www.cbs.dtu.dk/services/.
- ↑ "PSORT II Prediction". http://psort.hgc.jp/form2.html.
- ↑ "CELLO:Subcellular Localization Predictive System". http://cello.life.nctu.edu.tw/.
- ↑ 18.0 18.1 NCBI BLAST. BLAST AND PSI-Blast: generation of protein database search programs. [http://blast.ncbi.nlm.nih.gov/Blast.cgi ]
- ↑ "Bgee: Gene Expression Evolution. Entry on C8orf48 Homo Sapiens". BGEE. http://bgee.unil.ch/bgee/bgee?page=gene&action=summary&gene_id=ENSG000001647430.[yes|permanent dead link|dead link}}]
- ↑ 20.0 20.1 "EST Profile - Hs.104941". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.104941.
- ↑ 21.0 21.1 "101259684 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/101259684.
- ↑ 22.0 22.1 "47601584 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/47601584.
- ↑ 23.0 23.1 "MNT MAX network transcriptional repressor [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=ShowDetailView&TermToSearch=4335.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedStein_2008 - ↑ "Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core". Molecular and Cellular Biology 14 (3): 1786–95. 1994. doi:10.1128/mcb.14.3.1786. PMID 8114711.
- ↑ "ZKSCAN3 - Zinc finger protein with KRAB and SCAN domains 3 - Homo sapiens (Human) - ZKSCAN3 gene & protein". https://www.uniprot.org/uniprot/Q9BRR0.
- ↑ "T-box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration". Development 133 (24): 4945–55. 2006. doi:10.1242/dev.02694. PMID 17119020.
- ↑ "rProtein. Recombinant Protein Collection". http://www.rprotein.com/protein/ZNF219.[yes|permanent dead link|dead link}}]
- ↑ "www.genecards.org/cgi-bin/carddisp.pl?gene=KLF2". https://www.genecards.org/cgi-bin/carddisp.pl?gene=KLF2.
- ↑ "DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription". European Journal of Biochemistry 233 (1): 73–82. 1995. doi:10.1111/j.1432-1033.1995.073_1.x. PMID 7588776.
- ↑ "www.genecards.org/cgi-bin/carddisp.pl?gene=LRRFIP1". https://www.genecards.org/cgi-bin/carddisp.pl?gene=LRRFIP1.
- ↑ "E2F1 E2F transcription factor 1 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/1869.
- ↑ "KLF7 Kruppel-like factor 7 (ubiquitous) [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/8609.
- ↑ "CEBPE CCAAT/enhancer binding protein epsilon [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/1053.
- ↑ "POZ domain transcription factor, FBI-1, represses transcription of ADH5/FDH by interacting with the zinc finger and interfering with DNA binding activity of Sp1" (in en). The Journal of Biological Chemistry 277 (30): 26761–8. July 2002. doi:10.1074/jbc.M202078200. PMID 12004059.
- ↑ "OLIG2 oligodendrocyte lineage transcription factor 2 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/10215.
- ↑ "www.genecards.org/cgi-bin/carddisp.pl?gene=KLF3". https://www.genecards.org/cgi-bin/carddisp.pl?gene=KLF3.
 |