Biology:ChrB RNA motif
| chrB-b | |
|---|---|
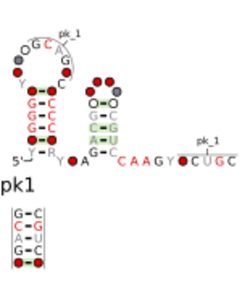 Consensus secondary structure and sequence conservation of chrB-b RNA | |
| Identifiers | |
| Symbol | chrB-b |
| Rfam | RF03086 |
| Other data | |
| RNA type | Cis-reg |
| SO | 0005836 |
| PDB structures | PDBe |
The chrB-a RNA motif and chrB-b RNA motif refer to a related, conserved RNA structure that was discovered by bioinformatics.[1] The structures of these motifs are similar, and some genomic locations are predicted to exhibit both motifs. The chrB-b motif has an extra pseudoknot that is not consistently found in chrB-a examples. It was proposed that the two motifs could be unified into one common structure, with additional information.[1]
Both motifs are found in Alphaproteobacteria and likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes. Additionally, the Shine-Dalgarno sequence of the genes is a part of the conserved secondary structure of the chrB motifs. Thus, a stabilization of this secondary structure is expected to reduce gene expression.
The genes apparently regulated by these RNAs share a relationship with each other in that they are involved in resistance to toxic levels of chromate:
- Many of these genes are classified as the gene chrB, which was found to improve the resistance of Ochrobactrum tritici to chromate.[2]
- Genes classified as chrA are also common.[2]
- Genes that encode the enzyme superoxide dismutase are also common, and this gene also has an established role in chromate resistance.[2]
- Rarely regulated genes with and without a relationship to chromate were also discussed.[1]
Additionally, a chrB gene regulated by a chrB-a RNA in Sinorhizobium meliloti strain 1021 is highly expressed in bacteria growing on plants.[1]
References
- ↑ 1.0 1.1 1.2 1.3 "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
- ↑ 2.0 2.1 2.2 "The chromate-inducible chrBACF operon from the transposable element TnOtChr confers resistance to chromium(VI) and superoxide". J. Bacteriol. 190 (21): 6996–7003. November 2008. doi:10.1128/JB.00289-08. PMID 18776016.
 |

