Biology:Comet assay
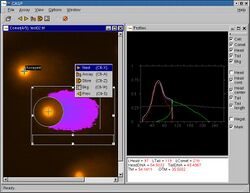
The single cell gel electrophoresis assay (SCGE, also known as comet assay) is an uncomplicated and sensitive technique for the detection of DNA damage at the level of the individual eukaryotic cell. It was first developed by Östling & Johansson in 1984 and later modified by Singh et al. in 1988.[1] It has since increased in popularity as a standard technique for evaluation of DNA damage/repair, biomonitoring and genotoxicity testing. It involves the encapsulation of cells in a low-melting-point agarose suspension, lysis of the cells in neutral or alkaline (pH>13) conditions, and electrophoresis of the suspended lysed cells. The term "comet" refers to the pattern of DNA migration through the electrophoresis gel, which often resembles a comet.[2][3]
The comet assay (single-cell gel electrophoresis) is a simple method for measuring deoxyribonucleic acid (DNA) strand breaks in eukaryotic cells. Cells embedded in agarose on a microscope slide are lysed with detergent and high salt to form nucleoids containing supercoiled loops of DNA linked to the nuclear matrix. Electrophoresis at high pH results in structures resembling comets, observed by fluorescence microscopy; the intensity of the comet tail relative to the head reflects the number of DNA breaks. The likely basis for this is that loops containing a break lose their supercoiling and become free to extend toward the anode. This is followed by visual analysis with staining of DNA and calculating fluorescence to determine the extent of DNA damage. This can be performed by manual scoring or automatically by imaging software.[4][5]
Procedure
Encapsulation
A sample of cells, either derived from an in vitro cell culture or from an in vivo test subject is dispersed into individual cells and suspended in molten low-melting-point agarose at 37 °C. This mono-suspension is cast on a microscope slide. A glass cover slip is held at an angle and the mono-suspension applied to the point of contact between the coverslip and the slide. As the coverslip is lowered onto the slide the molten agarose spreads to form a thin layer. The agarose is gelled at 4 °C and the coverslip removed.
The agarose forms a matrix of carbohydrate fibres that encapsulate the cells, anchoring them in place. The agarose is considered to be osmotic-neutral, therefore solutions can penetrate the gel and affect the cells without cells shifting position.
In an in vitro study the cells would be exposed to a test agent – typically UV light, ionising radiation, or a genotoxic chemical – to induce DNA damage in the encapsulated cells. For calibration, hydrogen peroxide is usually used to provide a standardized level of DNA damage.
Lysis
The slides are then immersed in a solution that cause the cells to lyse. The lysis solution often used in the comet assay consists of a highly concentrated aqueous salt (often, common table salt can be used) and a detergent (such as Triton X-100 or sarcosinate). The pH of the lysis solution can be adjusted (usually between neutral and alkaline pH) depending upon the type of damage the researcher is investigating.
The aqueous salt disrupts proteins and their bonding patterns within the cell as well as disrupting the RNA content of the cell. The detergent dissolves the cellular membranes. Through the action of the lysis solution the cells are destroyed. All proteins, RNA, membranes and cytoplasmic and nucleoplasmic constituents are disrupted and diffuse into the agarose matrix. Only the DNA of the cell remains, and unravels to fill the cavity in the agarose that the whole cell formerly filled. This structure is called nucleoid (a general term for a structure in which DNA is concentrated).
Electrophoresis
After lysis of the cells (typically 1 to 2 hours at 4 °C) the slides are washed in distilled water to remove all salts and immersed in a second solution – an electrophoresis solution. Again this solution can have its pH adjusted depending upon the type of damage that is being investigated.
The slides are left for ~20 minutes in the electrophoresis solution prior to an electric field being applied. In alkaline conditions the DNA double helix is denatured and the nucleoid becomes single stranded.
An electric field is applied (typically 1 V/cm) for ~20 minutes. The slides are then neutralised to pH 7, stained with a DNA-specific fluorescent stain and analysed using a microscope with an attached CCD (charge-coupled device – essentially a digital camera) that is connected to a computer with image analysis software.
Background
The concept underlying the SCGE assay is that undamaged DNA retains a highly organized association with matrix proteins in the nucleus. When damaged, this organization is disrupted. The individual strands of DNA lose their compact structure and relax, expanding out of the cavity into the agarose. When the electric field is applied the DNA, which has an overall negative charge, is drawn towards the positively charged anode. Undamaged DNA strands are too large and do not leave the cavity, whereas the smaller the fragments, the farther they are free to move in a given period of time. Therefore, the amount of DNA that leaves the cavity is a measure of the amount of DNA damage in the cell.
The image analysis measures the overall intensity of the fluorescence for the whole nucleoid and the fluorescence of the migrated DNA and compares the two signals. The stronger the signal from the migrated DNA the more damage there is present. The overall structure resembles a comet (hence "comet assay") with a circular head corresponding to the undamaged DNA that remains in the cavity and a tail of damaged DNA. The brighter and longer the tail, the higher the level of damage.
The comet assay is a versatile technique for detecting damage and with adjustments to the protocol can be used to quantify the presence of a wide variety of DNA altering lesions (damage). The damage usually detected are single strand breaks and double strand breaks. It is sometimes stated[6][7] that alkaline conditions and complete denaturating of the DNA is necessary to detect single strand breaks. However this is not true, both single- and double strand breaks are also detected in neutral conditions.[8][9][10][11] In alkaline conditions, however, additional DNA structures are detected as DNA damage: AP sites (abasic sites missing either a pyrimidine or purine nucleotide) and sites where excision repair is taking place.[12][13]
The comet assay is an extremely sensitive DNA damage assay. This sensitivity needs to be handled carefully as it is also vulnerable to physical changes which can affect the reproducibility of results. Essentially, anything that can cause DNA damage or denaturation except the factor(s) being researched is to be avoided.[14] The most common form of the assay is the alkaline version although there is as yet no definitive alkaline assay protocol. Due to its simple and inexpensive setup, it can be used in conditions where more complex assays are not available.
Applications
These include genotoxicity testing, human biomonitoring and molecular epidemiology, ecogenotoxicology, as well as fundamental research in DNA damage and repair.[15] For example, Swain and Rao, using the comet assay[16] reported marked increases in several types of DNA damages in rat brain neurons and astrocytes during aging, including single-strand breaks, double-strand breaks and modified bases (8-OHdG and uracil).
Sperm DNA fragmentation
A comet assay can determine the degree of DNA fragmentation in sperm cells. The degree of DNA fragmentation has been associated with outcomes of in vitro fertilization.[17][18]
The comet has been modified for use with sperm cells as a tool for male infertility diagnosis [19][20][21]
To break down these tightly bound protamine proteins in order to use the comet for sperm, additional steps in the de-condensation protocol are required.[20]
References
- ↑ Singh, Narendra P.; McCoy, Michael T.; Tice, Raymond R.; Schneider, Edward L. (1988-03-01). "A simple technique for quantitation of low levels of DNA damage in individual cells" (in en). Experimental Cell Research 175 (1): 184–191. doi:10.1016/0014-4827(88)90265-0. ISSN 0014-4827. PMID 3345800. https://dx.doi.org/10.1016/0014-4827%2888%2990265-0.
- ↑ Tice, R. R. (2000). "Single Cell Gel/Comet Assay: Guidelines for in vitro and in vivo Genetic Toxicology Testing". Environmental and Molecular Mutagenesis 35 (3): 206–21. doi:10.1002/(SICI)1098-2280(2000)35:3<206::AID-EM8>3.0.CO;2-J. PMID 10737956.
- ↑ Nandhakumar, S; Parasuraman, S; Shanmugam, M M; Rao, K R; Chand, P; Bhat, B V (2011). "Evaluation of DNA damage using single-cell gel electrophoresis (Comet Assay)". J Pharmacol Pharmacother 2 (2): 107–11. doi:10.4103/0976-500X.81903. PMID 21772771.
- ↑ Collins, A. R. (March 2004). "The comet assay for DNA damage and repair: principles, applications, and limitations". Mol. Biotechnol. 26 (3): 249–61. doi:10.1385/MB:26:3:249. PMID 15004294. https://zenodo.org/record/975797.
- ↑ Nandhakumar, S; Parasuraman, S; Shanmugam, MM; Rao, KR; Chand, P; Bhat, BV (Apr 2011). "Evaluation of DNA damage using single-cell gel electrophoresis (Comet Assay).". J Pharmacol Pharmacother 2 (2): 107–11. doi:10.4103/0976-500X.81903. PMID 21772771. PMC 3127337. http://www.jpharmacol.com/text.asp?2011/2/2/107/8190. Retrieved 2013-12-05.
- ↑ Pu, Xinzhu; Wang, Zemin; Klaunig, James E. (2015-08-06). "Alkaline Comet Assay for Assessing DNA Damage in Individual Cells". Current Protocols in Toxicology 65: 3.12.1–11. doi:10.1002/0471140856.tx0312s65. ISBN 978-0471140856. ISSN 1934-9262. PMID 26250399.
- ↑ Møller, Peter (April 2006). "The alkaline comet assay: towards validation in biomonitoring of DNA damaging exposures". Basic & Clinical Pharmacology & Toxicology 98 (4): 336–345. doi:10.1111/j.1742-7843.2006.pto_167.x. ISSN 1742-7835. PMID 16623855.
- ↑ Belyaev, Igor Y; Eriksson, Stefan; Nygren, Jonas; Torudd, Jesper; Harms-Ringdahl, Mats (1999-08-05). "Effects of ethidium bromide on DNA loop organisation in human lymphocytes measured by anomalous viscosity time dependence and single cell gel electrophoresis". Biochimica et Biophysica Acta (BBA) - General Subjects 1428 (2–3): 348–356. doi:10.1016/S0304-4165(99)00076-8. ISSN 0304-4165. PMID 10434054.
- ↑ Olive, P. L.; Banáth, J. P.; Durand, R. E. (April 1990). "Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the "comet" assay". Radiation Research 122 (1): 86–94. doi:10.2307/3577587. ISSN 0033-7587. PMID 2320728. Bibcode: 1990RadR..122...86O.
- ↑ Powell, S.; McMillan, T.J. (1990-10-01). "DNA damage and repair following treatment with ionizing radiation" (in en). Radiotherapy and Oncology 19 (2): 95–108. doi:10.1016/0167-8140(90)90123-E. ISSN 0167-8140. PMID 2255771. http://www.thegreenjournal.com/article/0167-8140(90)90123-E/abstract.
- ↑ Wojewódzka, Maria; Buraczewska, Iwona; Kruszewski, Marcin (2002-06-27). "A modified neutral comet assay: elimination of lysis at high temperature and validation of the assay with anti-single-stranded DNA antibody". Mutation Research 518 (1): 9–20. doi:10.1016/s1383-5718(02)00070-0. ISSN 0027-5107. PMID 12063063.
- ↑ Collins, Andrew R (1997-04-29). "The comet assay: what can it really tell us?". Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 375 (2): 183–193. doi:10.1016/S0027-5107(97)00013-4. ISSN 0027-5107. PMID 9202728.
- ↑ Gedik, C. M.; Ewen, S. W.; Collins, A. R. (September 1992). "Single-cell gel electrophoresis applied to the analysis of UV-C damage and its repair in human cells". International Journal of Radiation Biology 62 (3): 313–320. doi:10.1080/09553009214552161. ISSN 0955-3002. PMID 1356133.
- ↑ Theoretical and practical limitations to the assay are discussed e.g. in Klaude et al. (1996) and Collins et al. (1997).
- ↑ https://www2.le.ac.uk/departments/csmm/.../SCG%20Electrophoresis.pdf[yes|permanent dead link|dead link}}]
- ↑ Swain, U; Subba Rao, K (Aug 2011). "Study of DNA damage via the comet assay and base excision repair activities in rat brain neurons and astrocytes during aging". Mech Ageing Dev. 132 (8–9): 374–81. doi:10.1016/j.mad.2011.04.012. PMID 21600238. S2CID 22466782
- ↑ "Clinical significance of sperm DNA damage in assisted reproduction outcome". Hum Reprod 25 (7): 1594–1608. May 2010. doi:10.1093/humrep/deq103. PMID 20447937.
- ↑ Nagrajappa; Ganguly, Anutosh; Goswami, Usha (2006). "DNA damage in male gonad cells of Green mussel (Perna viridis) upon exposure to tobacco products.". Ecotoxicology 15 (4): 365–369. doi:10.1007/s10646-006-0077-1. PMID 16673160.
- ↑ Hughes CM, Lewis SEM, McKelvey-Martin V, Thompson W. Reproducibility of human sperm DNA measurements using a single cell gel electrophoresis assay. Mutation Research 1997 374:261-268.
- ↑ 20.0 20.1 Donnelly, ET; McClure, N; Lewis, SEM (1999). "The effects of ascorbate and @-tocopherol supplementation in vitro on DNA integrity and hydrogen peroxide-induced DNA damage in human spermatozoa". Mutagenesis 14 (5): 505–511. doi:10.1093/mutage/14.5.505. PMID 10473655.
- ↑ Ribas-Maynou Jordi, Agustı´n Garcı´a-Peiro´ , Alba Fernandez-Encinas, Maria Jose´ Amengual and Benet Jordi ,Double Stranded Sperm DNA Breaks, Measured by Comet Assay, Are Associated with Unexplained Recurrent Miscarriage in Couples without a Female Factor
Further reading
- Dhawan & Anderson (2009): The Comet Assay in Toxicology. doi:10.1039/9781847559746
- Avishai, Nanthawan; Rabinowitz, Claudette; Moiseeva, Elisabeth; Rinkevich, Baruch (2002). "Genotoxicity of the Kishon River, Israel: the application of an in vitro cellular assay". Mutation Research 518 (1): 21–37. doi:10.1016/S1383-5718(02)00069-4. PMID 12063064.
- Collins, A.R.; Dobson, V.L.; Dusinska, M.; Kennedy, G.; Stetina, R. (1997). "The comet assay: what can it really tell us?". Mutation Research 375 (2): 183–193. doi:10.1016/S0027-5107(97)00013-4. PMID 9202728.
- Klaude, M.; Eriksson, S.; Nygren, J.; Ahnstrom, G. (1996). "The comet assay: mechanisms and technical considerations". Mutation Research 363 (2): 89–96. doi:10.1016/0921-8777(95)00063-1. PMID 8676929.
- McKelvey-Martin, Valerie J.; Ho, Edwin T.; McKeown, Stephanie R.; Johnston, S. Robin; McCarthy, Patsy J.; Rajab, Nor Fasilah; Downes, C. Stephen (1993). "Emerging applications of the single cell gel electrophoresis (Comet) assay. I. Management of invasive transitional cell human bladder carcinoma. II. Fluorescent in situ hybridization Comets for the identification of damaged and repaired DNA sequences in individual cells". Mutagenesis 13 (1): 1–8. doi:10.1093/mutage/13.1.1. PMID 9491387.
- Olive, P.L.; Wlodek, D.; Banath, J.P. (1991). "DNA double-strand breaks measured in individual cells subjected to gel electrophoresis". Cancer Research 51 (17): 4671–4676. PMID 1873812. http://cancerres.aacrjournals.org/cgi/reprint/51/17/4671.pdf.
- Rojas, E.; Lopez, M.C.; Valverde, M. (1999). "Single cell gel electrophoresis: methodology and applications". Journal of Chromatography B 722 (1–2): 225–254. doi:10.1016/S0378-4347(98)00313-2. PMID 10068143.
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. (1988). "A simple technique for quantitation of low levels of DNA damage in individual cells". Experimental Cell Research 175 (1): 184–191. doi:10.1016/0014-4827(88)90265-0. PMID 3345800. https://zenodo.org/record/1253874.
- Tice, R.R.; Agurell, E.; Anderson, D.; Burlinson, B.; Hartmann, A.; Kobayashi, H.; Miyamae, Y.; Rojas, E. et al. (2000). "Single Cell Gel/Comet Assay: Guidelines for In Vitro and In Vivo Genetic Toxicology Testing". Environmental and Molecular Mutagenesis 35 (3): 206–221. doi:10.1002/(SICI)1098-2280(2000)35:3<206::AID-EM8>3.0.CO;2-J. PMID 10737956.
 |
