Biology:FAM149B1
The Family with sequence similarity 149 member B1 is an uncharacterized protein[1] encoded by the human FAM149B1 gene, with one alias KIAA0974.[2][3] The protein resides in the nucleus of the cell. The predicted secondary structure of the gene contains multiple alpha-helices, with a few beta-sheet structures. The gene is conserved in mammals, birds, reptiles, fish, and some invertebrates. The protein encoded by this gene contains a DUF3719 protein domain, which is conserved across its orthologues.[3] The protein is expressed at slightly below average levels in most human tissue types, with high expression in brain, kidney, and testes tissues, while showing relatively low expression levels in pancreas tissues.[4][5]
Gene
This gene has a possible 14 exons. It is located on the forward strand of chromosome 10 at 10q22.2 on the positive strand.[6] The total span of the gene, including 5' and 3' UTR, is 3149 base pairs. The gene is flanked on the left by NUDT13 (nudix hydrolase 13) and on the right by DNAJC9-AS1 (DNAJC9 antisense RNA 1).

Isoforms
The FAM149B1 protein has a possible 10 isoforms, which are determined through alternative splicing of the gene.

| Isoform Name | Accession | Exons | Length (bp) |
| Primary Transcript | NM_173348.1 | All (14) | 3149 |
| X1 | XM_005269744.2 | All (14) | 3108 |
| X2 | XM_011539737.2 | 13 | 2935 |
| X3 | XM_005269745.2 | 13 | 3006 |
| X4 | XM_017016164.1 | 12 | 2810 |
| X5 | XM_017016165.1 | 11 | 2779 |
| X6* | XM_017016166.1 | 9 | 2816 |
| X6* | XM_005269747.3 | 9 | 2923 |
| X7 | XM_017016167.1 | 9 | 1485 |
| X8 | XM_011539740.2 | 9 | 1447 |
Protein
General properties
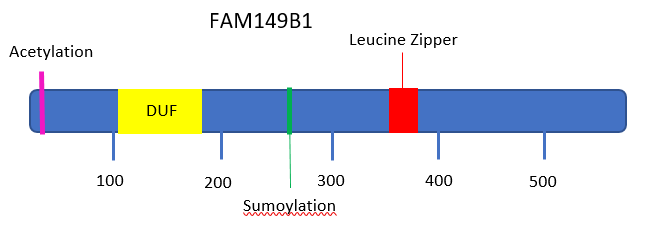
The primary protein encoded by the FAM149B1 gene is 583 amino acids in length and has a molecular weight of 64 kDal. The protein contains a conserved protein domain, DUF3719[8][6] located at the amino acids 115–179. The isoelectric point of the protein before post-translational modifications is 6.3,[9] and this isoelectric point is relatively conserved in the protein's isoforms, especially in those with the most similar composition of exons. This protein is considered serine rich, in that it expresses a higher serine composition relative to the composition of other human proteins.[10][11] This high serine composition is also seen in the gene's orthologues.
Splice variants
The splice variants of the protein demonstrate some shared qualities of the protein that is translated from the primary transcript. Because each isoform is a different length and contains various combinations of the available exons, there are variances in the isoelectric point and molecular weight. The isoforms closest to the weight and exon composition to the primary transcript generally share these characteristics. The protein isoforms missing the conserved DUF3719 domain are isoforms X5 and X6 because this domain is contained between exons 3–6.
| Isoform Name | Accession | Molecular Weight (kDal) | Length (aa) | Isoelectric point |
| Primary Transcript | NP_775483.1 | 64 | 582 | 6.3 |
| X1 | XP_005269801.1 | 63.7 | 574 | 6.3 |
| X2 | XP_011538039.1 | 62.6 | 560 | 7.5 |
| X3 | XP_005269802.1 | 59.8 | 540 | 6.4 |
| X4 | XP_016871653.1 | 57.8 | 518 | 7.7 |
| X5 | XP_016871654.1 | 53 | 476 | 6.8 |
| X6* | XP_016871655.1 | 46.6 | 419 | 7.5 |
| X6* | XP_005269804.1 | 46.6 | 419 | 7.5 |
| X7 | XP_016871656.1 | 41 | 368 | 5.1 |
| X8 | XP_011538042.1 | 38 | 348 | 5.2 |
Structure
There is a negative charge cluster from amino acids 212 to 239. Negative charge clusters often coordinate calcium, or magnesium or zinc ions, mannose-binding protein, or aminopeptidase.[12] The protein contains no positive or mixed charge clusters. The secondary structure of the protein is predicted to be a combination of mostly alpha-helices with a few predicted beta-sheet structures.

Subcellular localization

The subcellular location of the protein is the nucleus.[13] There is a leucine zipper pattern in the protein beginning at amino acid 347.[14]
Post-translational modifications
Acetylation
The third amino acid in the protein sequence, serine, is predicted to be acetylated.[15]
Phosphorylation
There are multiple predicted phosphorylation sites on various serine, tyrosine, and threonine amino acids are predicted for this protein sequence.[16] The conserved DUF3719 domain contains 7 predicted phosphorylation sites.
Sumoylation
One predicted sumoylation site was identified in the protein sequence at K267.[17]
Expression
Overall in the human body, this gene is expressed at levels slightly below the average human gene expression level.[18] The protein is expressed in most cell types of the human body.[19] Most experimentation shows a higher expression of this protein in kidney, testes, and brain tissues, with very low expression seen in pancreas tissues.[4][5] The gene is expressed at lower levels than its normal expression in most cancerous tissues. The gene is also seen to be expressed most highly in fetal and infantile tissues.[20]
DNA microarray data

DNA microarray analysis experiments show expression patterns of FAM149B1 compared to multiple other genes in a sample. FAM149B1 is shown to be at a lower expression level than most other genes in a multiple myeloma cell line and was shown to increase to close to average gene expression levels after the beta-catenin was depleted from the sample.[21]

FAM149B1 expression was also shown to decrease to lower than average gene expression levels in an ovarian cancer cell line after the use of an anticancer drug named NSC319726.[13]
Transcriptional regulation
The gene has nine different identified promoter regions, which correlate to the various isoforms of the gene. The promoter for the primary transcript of the gene has binding sites for a variety of different transcription factors.
Interacting proteins
Current data supports the FAM149B1 protein interactions with 6 different proteins.
One protein was determined to be an interacting protein with FAM149B1 through affinity chromatography techniques.
- The TBC1D32 protein has a role in many biological processes including determination of left/right symmetry, embryonic digit morphogenesis, heart development, lens development in camera-type eye, non-motile cilium assembly, protein localization to cilium, retinal pigment epithelium development, and smoothened signaling pathway involved in dorsal/ventral neural tube patterning. This protein is classified as a developmental protein.[22]
The other five proteins that have been predicted to interact with FAM149B1 protein were found through the process of textmining.
- The human ABHD8 (Alpha/Beta Hydrolase Domain-Containing Protein 8) protein is a hydrolase protein found in the extracellular exosome.[23]
- METTL16 (Methyltransferase 10 Domain Containing) protein is a methyltransferase found in the nucleus and cytosol of the cell.[24] This protein was experimentally determined to interact with YARS protein (tyrosyl-tRNA synthetase), which catalyzes attachment of tyrosine to tRNA, and MEPCE (Methylphosphate Capping Enzyme) which adds a methylphosphate cap at the 5-end of 7SK snRNA.
- SLC6A17 (Solute Carrier Family 6 Member 17) is a sodium-dependent vesicular transporter that is selective for proline, glycine, leucine, and alanine, and is not chloride-dependent as are most other transporters in its family.[25]
- The TM2D1 (Beta-Amyloid-Binding Protein) gene is thought to participate in beta-amyloid-induced apoptosis with its interaction with beta-APP42, and also has G-protein coupled receptor activity. This protein is found mainly in the nucleus and plasma membrane of the cell.[26]
- The DNAJc9 (DnaJ Heat Shock Protein Family (Hsp40) Member C9) protein may play a role as co-chaperone of the Hsp70 family proteins HSPA1A, HSPA1B and HSPA8. This protein is found extracellularly, and in the nucleus, plasma membrane, and cytosol.
Homology/Evolution
Paralog
There is one known paralog, FAM149A.[27] It is located on the human chromosome 4 at 4q35.1. The function of the protein encoded by this gene is not well understood, but it also contains the DUF3719 protein domain. The protein translated by this gene shares a 21.2% identity[28] with the FAM149B1 protein. The protein sequence is 482 amino acids in length.
Orthologs


This gene has orthologues across mammals, birds, reptiles, fish, and some invertebrates.[3] There is a high conservation in mammals, moderate conservation in many of the other vertebrate orthologues, and a low conservation in its few invertebrate orthologues.[29][28]
| Genus species | Common Name | Time of Divergence (MYA)[30] | Accession Number | Length (aa) | Identity[28] | |
|---|---|---|---|---|---|---|
| 1 | Homo sapiens | Human | - | NP_775483.1 | 582 | 100% |
| 2 | Pongo abelii | Sumatran orangutan | 15.76 | XP_009243761.1 | 587 | 93.0% |
| 3 | Papio anubis | Baboon | 29.4 | XP_003903829.1 | 582 | 93.6% |
| 4 | Mus musculus | Mouse | 90 | XP_006518391.1 | 544 | 73.5% |
| 5 | Bos mutus | Domestic Yak | 96 | XP_005910201.1 | 584 | 86.0% |
| 6 | Orcinus orca | Killer whale, Orca | 96 | XP_004273176.1 | 585 | 87.0% |
| 7 | Ailuropoda melanoleuca | Giant Panda | 96 | XP_011224744.1 | 590 | 82.7% |
| 8 | Orycteropus afer afer | Aardvark | 105 | XP_007938812.1 | 583 | 84.0% |
| 9 | Monodelphis domestica | Short-Tailed Opossum | 159 | XP_007478430.1 | 587 | 73.5% |
| 10 | Sarcophilus harrisii | Tasmanian Devil | 159 | XP_012396086.1 | 588 | 72.0% |
| 11 | Ornithorhynchus anatinus | Platypus | 177 | XP_007658720.1 | 506 | 48.1% |
| 12 | Gallus gallus | Chicken | 312 | XP_004942035.1 | 602 | 50.4% |
| 13 | Lepidothrix coronata | Blue-crowned manakin | 312 | XP_017688171.1 | 576 | 47.5% |
| 14 | Haliaeetus albicilla | White-tailed eagle | 312 | XP_009911204.1 | 589 | 49.4% |
| 15 | Falco peregrinus | Peregrine falcon | 312 | XP_005235226.1 | 597 | 49.2% |
| 16 | Chrysemys picta bellii | Western painted turtle | 312 | XP_008169104.1 | 596 | 56.1% |
| 17 | Pelodiscus sinensis | Chinese softshell turtle | 312 | XP_014433498.1 | 487 | 47.1% |
| 18 | Alligator mississippiensis | American alligator | 312 | XP_014464842.1 | 596 | 55.0% |
| 19 | Xenopus tropicalis | Western clawed frog | 352 | NP_001278638.1 | 561 | 39.8% |
| 20 | Danio rerio | Zebra fish | 435 | NP_001074134.1 | 644 | 37.7% |
| 21 | Lepisosteus oculatus | Spotted gar | 435 | XP_015202055.1 | 647 | 37.9% |
| 22 | Oreochromis niloticus | Nile tilapia | 435 | XP_005474333.1 | 683 | 34.3% |
| 23 | Callorhinchus milii | Australian ghostshark | 473 | XP_007897395.1 | 638 | 36.8% |
| 24 | Ciona intestinalis | Sea squirt | 676 | XP_002129894.1 | 807 | 24.5% |
| 25 | Aplysia californica | California sea slug | 797 | XP_012945921.1 | 312 | 16.9% |
Clinical significance
While the gene is largely not well understood by scientists, it is shown to be associated with a wide range of various cancerous tumors.[31][32]
The FAM149B1 gene is also included in a region of 11 genes that comprises one of 15 regions containing mutations associated with the African Pygmy phenotype.[33][34]
References
- ↑ "Homo sapiens family with sequence similarity 149 member B1 (FAM149B1), mRNA". https://www.ncbi.nlm.nih.gov/nuccore/NM_173348.1.
- ↑ "FAM149B1 Family with sequence similarity 149 member B1". https://www.ncbi.nlm.nih.gov/gene/317662.
- ↑ 3.0 3.1 3.2 "Gene: FAM149B1". http://useast.ensembl.org/Homo_sapiens/Gene/Summary?g=ENSG00000138286;r=10:73168166-73244504.
- ↑ 4.0 4.1 The Human Protein Atlas, FAM149B1 http://www.proteinatlas.org/ENSG00000138286-FAM149B1/tissue
- ↑ 5.0 5.1 NCBI, National Center for Biotechnology Information, H. sapiens FAM149B1 ETS https://www.ncbi.nlm.nih.gov/gene/317662
- ↑ 6.0 6.1 "FAM149B1 Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM149B1&keywords=fam149b1.
- ↑ "FAM149B1 family with sequence similarity 149 member B1 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/317662.
- ↑ "Protein of unknown function DUF3719 (IPR022194)". http://www.ebi.ac.uk/interpro/entry/IPR022194.
- ↑ PI, Biology Workbench. Program by Dr. Luca Toldo, developed at http://www.embl-heidelberg.de. Changed by Bjoern Kindler to print also the lowest found net charge. Available at EMBL WWW Gateway to Isoelectric Point Service {{cite web |url=http://www.embl-heidelberg.de/cgi/pi-wrapper.pl |title=Archived copy |access-date=2014-05-10 |url-status=dead |archive-url=https://web.archive.org/web/20081026062821/http://www.embl-heidelberg.de/cgi/pi-wrapper.pl |archive-date=2008-10-26 }}
- ↑ AASTATS, Biology Workbench. by Jack Kramer, 1990.
- ↑ SAPS, Biology Workbench. Algorithm: Brendel, V., Bucher, P., Nourbakhsh, I.R., Blaisdell, B.E. & Karlin, S. (1992) "Methods and algorithms for statistical analysis of protein sequences" Proc. Natl. Acad. Sci. U.S.A. 89, 2002-2006. Program: Volker Brendel, Department of Mathematics, Stanford University, Stanford CA 94305, U.S.A.
- ↑ Zhu, Z. Y.; Karlin, S. (1996-08-06). "Clusters of charged residues in protein three-dimensional structures" (in en). Proceedings of the National Academy of Sciences 93 (16): 8350–8355. doi:10.1073/pnas.93.16.8350. ISSN 0027-8424. PMID 8710874. Bibcode: 1996PNAS...93.8350Z.
- ↑ 13.0 13.1 13.2 13.3 "GDS4877 / 213463_s_at". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS4877:213463_s_at.
- ↑ "PSORT prediction tool". http://www.genscript.com/tools/psort.
- ↑ NetAcet: Prediction of N-terminal acetylation sites., Accessed through Expasy. Lars Kiemer, Jannick Dyrløv Bendtsen and Nikolaj Blom. Accepted in Bioinformatics, 2004.
- ↑ NetPhos: Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Proteomics: Jun;4(6):1633-49, review 2004.
- ↑ SUMOplot Analysis Program, Developed by Abgent, Copyright © 2013 -2017 http://www.abgent.com/sumoplot
- ↑ "FAM149B1 search results" (in en). http://pax-db.org/search?organism=&q=FAM149B1.
- ↑ NCBI GeoProfile, Homo sapiens FAM149B1, Profile GDS596 https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS596%3A213463_s_at
- ↑ Group, Schuler. "EST Profile - Hs.408577". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.408577.
- ↑ 21.0 21.1 "GDS3578 / 213463_s_at". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS3578:213463_s_at.
- ↑ Huttlin, E., Ting, L., Bruckner, R., Gebreab, F., Gygi, M., Szpyt, J., . . . Gygi, S. (2015). The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell, 162(2), 425-440. doi:10.1016/j.cell.2015.06.043
- ↑ "ABHD8_HUMAN". https://www.uniprot.org/uniprot/Q96I13#Q96I13-1.
- ↑ "METTL16 Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=METTL16.
- ↑ "SLC6A17 Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=SLC6A17.
- ↑ "TM2D1 Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=TM2D1.
- ↑ "Homo sapiens family with sequence similarity 149 member A (FAM149A), t - Nucleotide - NCBI". https://www.ncbi.nlm.nih.gov/nuccore/NM_001006655.3.
- ↑ 28.0 28.1 28.2 "ALIGN". http://workbench.sdsc.edu/.
- ↑ "NCBI BLAST". https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins.
- ↑ "Time Tree". http://www.timetree.org/.
- ↑ Ikeda, A; Shimizu, T; Matsumoto, Y (19 September 2013). "Leptin receptor somatic mutations are frequent in HCV-infected cirrhotic liver and associated with haptocellular carcinoma". Gastroenterology 146 (1): 222–32. doi:10.1053/j.gastro.2013.09.025. PMID 24055508.
- ↑ Hadj-Hamou, NS; Lae, M; Almeida, A (24 April 2012). "A transcriptome signature of endothelial lymphatic cells coexists with the chronic oxidative stress signature in radiation-induced post-radiotherapy breast angiosarcomas.". Carcinogenesis 33 (7): 1399–405. doi:10.1093/carcin/bgs155. PMID 22532251.
- ↑ Detection of Convergent Genome-Wide Signals of Adaptation to Tropical Forests in Humans. (Research Article) Amorim, Carlos Eduardo G. ; Daub, Josephine T. ; Salzano, Francisco M. ; Foll, Matthieu ; Excoffier, Laurent PLoS ONE, April 7, 2015, Vol.10(4), p.e0121557 [Peer Reviewed Journal]
- ↑ Adaptive evolution of loci covarying with the human African Pygmy phenotype Mendizabal, Isabel ; Marigorta, Urko ; Lao, Oscar ; Comas, David Human Genetics, 2012, Vol.131(8), pp.1305-1317 [Peer Reviewed Journal]
 |
