Biology:Flow-FISH
Flow-FISH (fluorescence in-situ hybridization) is a cytogenetic technique to quantify the copy number of RNA or specific repetitive elements in genomic DNA of whole cell populations via the combination of flow cytometry with cytogenetic fluorescent in situ hybridization staining protocols.[1][2][3]
Flow-FISH is most commonly used to quantify the length of telomeres, which are stretches of repetitious DNA (hexameric TTAGGG repeats) at the distal ends of chromosomes[4] in human white blood cells, and a semi-automated method for doing so was published in Nature Protocols.[1] Telomere length in white blood cells has been a subject of interest because telomere length in these cell types (and also of other somatic tissues) declines gradually over the human lifespan, resulting in cell senescence, apoptosis,[5] or transformation.[6] This decline has been shown to be a surrogate marker for the concomitant decline in the telomere length of the hematopoietic stem cell pool, with the granulocyte lineage giving the best indication, presumably due to the absence of a long lived memory subtype and comparatively rapid turnover of these cells.[7]
Flow-FISH is also suitable for the concomitant detection of RNA and protein.[2] This allows for the identification of cells that not only express a gene, but also translate it into protein. This type of Flow-FISH has been used to study latent infection of viruses such as HIV-1 and EBV,[8][9] but also to track single cell gene expression and translation into protein.[2][10]
Q-FISH to flow-FISH
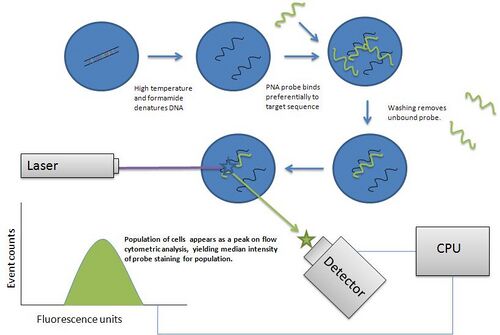
Flow-FISH was first published in 1998 by Rufer et al.[11] as a modification of another technique for analyzing telomere length, Q-FISH, that employs peptide nucleic acid probes[12] of a 3'-CCCTAACCCTAACCCTAA-5' sequence labeled with a fluorescin fluorophore to stain telomeric repeats on prepared metaphase spreads of cells that have been treated with colcemid, hypotonic shock, and fixation to slides via methanol/acetic acid treatment[13] Images of the resultant fluorescent spots could then be analyzed via a specialized computer program to yield quantitative fluorescence values that can then be used to estimate actual telomere length. The fluorescence yielded by probe staining is considered to be quantitative because PNA binds preferentially to DNA at low ionic salt concentrations and in the presence of formamide, thus the DNA duplex may not reform once it has been melted and annealed to PNA probe, allowing the probe to saturate its target repeat sequence (as it is not displaced from the target DNA by competing anti sense DNA on the complementary strand), thus yielding a reliable and quantifiable readout of the frequency of PNA probe target at a given chromosomal site after washing away of unbound probe.[13]
Innovation
Unlike Q-FISH, Flow-FISH utilizes the quantitative properties of telomere specific PNA probe retention to quantify median fluorescence in a population of cells, via the use of a flow cytometer, instead of a fluorescence microscope.[14] The primary advantage of this technique is that it eliminates the time required in Q-FISH to prepare metaphase spreads of cells of interest, and that flow cytometric analysis is also considerably faster than the methods required to acquire and analyze Q-FISH prepared slides. Flow-FISH thus allows for a higher throughput analysis of telomere length in blood leukocytes, which are a readily available form of human tissue sample. The most recent versions of the flow-FISH technique include an internal control population of cow thymocytes with a known telomere length detected by TRF or telomere restriction fragment analysis to which the fluorescence of a given unknown sample may be compared. Because cow thymocytes take up LDS751 dye to a lesser extent than their human counterparts, they may be reliably differentiated via plotting and gating the desired populations. Other cell types that have not in the past proven to be good candidates for flow-FISH can be analyzed via extraction of nuclei and performance of the technique on them directly.[15]

References
- ↑ 1.0 1.1 Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc 2006; 1:2365–2376.
- ↑ 2.0 2.1 2.2 Porichis, Filippos; Hart, Meghan G.; Griesbeck, Morgane; Everett, Holly L.; Hassan, Muska; Baxter, Amy E.; Lindqvist, Madelene; Miller, Sara M. et al. (December 2014). "High-throughput detection of miRNAs and gene-specific mRNA at the single-cell level by flow cytometry" (in en). Nature Communications 5 (1): 5641. doi:10.1038/ncomms6641. ISSN 2041-1723. PMID 25472703. Bibcode: 2014NatCo...5.5641P.
- ↑ Baxter, Amy E.; Niessl, Julia; Fromentin, Rémi; Richard, Jonathan; Porichis, Filippos; Massanella, Marta; Brassard, Nathalie; Alsahafi, Nirmin et al. (October 2017). "Multiparametric characterization of rare HIV-infected cells using an RNA-flow FISH technique" (in en). Nature Protocols 12 (10): 2029–2049. doi:10.1038/nprot.2017.079. ISSN 1750-2799. PMID 28880280.
- ↑ Moyzis, R.K. et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 85, 6622–6626 (1988).
- ↑ Harley, C.B., Futcher, A.B. & Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 (1990).
- ↑ Chang, S., Khoo, C.M., Naylor, M.L., Maser, R.S. & DePinho, R.A. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 17, 88–100 (2003).
- ↑ Rufer N, Brummendorf TH, Kolvraa S, et al. Telomere fluorescence measurements in granulocytes and T lymphocyte subsets point to a high turnover of hematopoietic stem cells and memory T cells in early childhood. J Exp Med 1999; 190:157–167.
- ↑ Grau-Expósito, Judith; Luque-Ballesteros, Laura; Navarro, Jordi; Curran, Adrian; Burgos, Joaquin; Ribera, Esteban; Torrella, Ariadna; Planas, Bibiana et al. (2019-08-19). Swanstrom, Ronald. ed. "Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations" (in en). PLOS Pathogens 15 (8): e1007991. doi:10.1371/journal.ppat.1007991. ISSN 1553-7374. PMID 31425551.
- ↑ Fournier, Benjamin; Boutboul, David; Bruneau, Julie; Miot, Charline; Boulanger, Cécile; Malphettes, Marion; Pellier, Isabelle; Dunogué, Bertrand et al. (2020-11-02). "Rapid identification and characterization of infected cells in blood during chronic active Epstein-Barr virus infection" (in en). Journal of Experimental Medicine 217 (11): e20192262. doi:10.1084/jem.20192262. ISSN 0022-1007. PMID 32812031.
- ↑ Nicolet, Benoit P.; Guislain, Aurelie; Wolkers, Monika C. (2017-01-15). "Combined Single-Cell Measurement of Cytokine mRNA and Protein Identifies T Cells with Persistent Effector Function" (in en). The Journal of Immunology 198 (2): 962–970. doi:10.4049/jimmunol.1601531. ISSN 0022-1767. PMID 27927969.
- ↑ Rufer, N., Dragowska, W., Thornbury, G., Roosnek, E. & Lansdorp, P.M. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nature Biotechnol. 16, 743–747 (1998).
- ↑ Egholm, M. et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 365, 566–568 (1993).
- ↑ 13.0 13.1 Lansdorp, P.M. et al. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5, 685–691 (1996).
- ↑ Baerlocher, G.M. & Lansdorp, P.M. Telomere length measurements in leukocyte subsets by automated multicolor flow FISH. Cytometry A 55, 1–6 (2003).
- ↑ Wieser, M. et al. Nuclear flow FISH: isolation of cell nuclei improves the determination of telomere lengths. Exp. Gerontol. 41, 230–235 (2006).
 |
