Biology:FuFi-1 RNA motif
| FuFi-1 | |
|---|---|
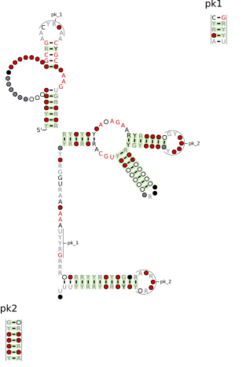 Consensus secondary structure and sequence conservation of FuFi-1 RNA | |
| Identifiers | |
| Symbol | FuFi-1 |
| Rfam | RF02986 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0001263 |
| PDB structures | PDBe |
The FuFi-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] Such RNA "motifs" are often the first step to elucidating the biological function of a novel RNA.[2] FuFi-1 motif RNAs are found in Bacillota AND Fusobacteriota.
FuFi-1 RNAs are sometimes located in the vicinity of genes that encode XRE-like proteins, an example of a helix-turn-helix structure. However, XRE-like proteins are very common in bacteria, and therefore it is unclear that this association represents an important biological connection. Also, while the XRE-like genes are often located nearby to the FuFi-1 RNAs, the RNAs are not positioned so that they could be consistently located in the 5' untranslated region of the genes. Therefore, FuFi-1 RNAs likely function in trans as small RNAs. Predicted Rho-independent transcription terminator hairpins occur on the 3' part of the FuFi-1 RNA motif, suggesting that they terminate transcripts containing the RNA motif. These transcription terminators were predicting using the RNie program.[3]
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
- ↑ "Challenges of ligand identification for the second wave of orphan riboswitch candidates". RNA Biol 15 (3): 377–390. March 2018. doi:10.1080/15476286.2017.1403002. PMID 29135333.
- ↑ "RNIE: genome-wide prediction of bacterial intrinsic terminators". Nucleic Acids Res. 39 (14): 5845–5852. August 2011. doi:10.1093/nar/gkr168. PMID 21478170.
 |

