Biology:Helix-turn-helix
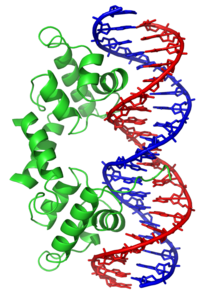
Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.[1]
Discovery
The discovery of the helix-turn-helix motif was based on similarities between several genes encoding transcription regulatory proteins from bacteriophage lambda and Escherichia coli: Cro, CAP, and λ repressor, which were found to share a common 20–25 amino acid sequence that facilitates DNA recognition.[2][3][4][5]
Function
The helix-turn-helix motif is a DNA-binding motif. The recognition and binding to DNA by helix-turn-helix proteins is done by the two α helices, one occupying the N-terminal end of the motif, the other at the C-terminus. In most cases, such as in the Cro repressor, the second helix contributes most to DNA recognition, and hence it is often called the "recognition helix". It binds to the major groove of DNA through a series of hydrogen bonds and various Van der Waals interactions with exposed bases. The other α helix stabilizes the interaction between protein and DNA, but does not play a particularly strong role in its recognition.[2] The recognition helix and its preceding helix always have the same relative orientation.[6]
Classification of helix-turn-helix motifs
Several attempts have been made to classify the helix-turn-helix motifs based on their structure and the spatial arrangement of their helices.[6][7][8] Some of the main types are described below.
Di-helical
The di-helical helix-turn-helix motif is the simplest helix-turn-helix motif. A fragment of Engrailed homeodomain encompassing only the two helices and the turn was found to be an ultrafast independently folding protein domain.[9]
Tri-helical
An example of this motif is found in the transcriptional activator Myb.[10]
Tetra-helical
The tetra-helical helix-turn-helix motif has an additional C-terminal helix compared to the tri-helical motifs. These include the LuxR-type DNA-binding HTH domain found in bacterial transcription factors and the helix-turn-helix motif found in the TetR repressors.[11] Multihelical versions with additional helices also occur.[12]
Winged helix-turn-helix
The winged helix-turn-helix (wHTH) motif is formed by a 3-helical bundle and a 3- or 4-strand beta-sheet (wing). The topology of helices and strands in the wHTH motifs may vary. In the transcription factor ETS wHTH folds into a helix-turn-helix motif on a four-stranded anti-parallel beta-sheet scaffold arranged in the order α1-β1-β2-α2-α3-β3-β4 where the third helix is the DNA recognition helix.[13][14]
Other modified helix-turn-helix motifs
Other derivatives of the helix-turn-helix motif include the DNA-binding domain found in MarR, a regulator of multiple antibiotic resistance, which forms a winged helix-turn-helix with an additional C-terminal alpha helix.[8][15]
See also
References
- ↑ "The helix-turn-helix DNA binding motif". The Journal of Biological Chemistry 264 (4): 1903–6. February 1989. doi:10.1016/S0021-9258(18)94115-3. PMID 2644244.
- ↑ 2.0 2.1 "Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor". Proceedings of the National Academy of Sciences of the United States of America 79 (5): 1428–32. March 1982. doi:10.1073/pnas.79.5.1428. PMID 6951187. Bibcode: 1982PNAS...79.1428M.
- ↑ "Structure of the cro repressor from bacteriophage lambda and its interaction with DNA". Nature 290 (5809): 754–8. April 1981. doi:10.1038/290754a0. PMID 6452580. Bibcode: 1981Natur.290..754A.
- ↑ "Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA". Nature 290 (5809): 744–9. April 1981. doi:10.1038/290744a0. PMID 6261152.
- ↑ "The operator-binding domain of lambda repressor: structure and DNA recognition". Nature 298 (5873): 443–7. July 1982. doi:10.1038/298443a0. PMID 7088190. Bibcode: 1982Natur.298..443P.
- ↑ 6.0 6.1 "Structural classification of HTH DNA-binding domains and protein-DNA interaction modes". Journal of Molecular Biology 262 (2): 294–313. September 1996. doi:10.1006/jmbi.1996.0514. PMID 8831795.
- ↑ "Classification of multi-helical DNA-binding domains and application to predict the DBD structures of sigma factor, LysR, OmpR/PhoB, CENP-B, Rapl, and Xy1S/Ada/AraC". FEBS Letters 372 (2–3): 215–21. September 1995. doi:10.1016/0014-5793(95)00988-L. PMID 7556672.
- ↑ 8.0 8.1 "The many faces of the helix-turn-helix domain: transcription regulation and beyond". FEMS Microbiology Reviews 29 (2): 231–62. April 2005. doi:10.1016/j.femsre.2004.12.008. PMID 15808743. https://zenodo.org/record/1258943.
- ↑ "The helix-turn-helix motif as an ultrafast independently folding domain: the pathway of folding of Engrailed homeodomain". Proceedings of the National Academy of Sciences of the United States of America 104 (22): 9272–7. May 2007. doi:10.1073/pnas.0703434104. PMID 17517666. Bibcode: 2007PNAS..104.9272R.
- ↑ "Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core". Proceedings of the National Academy of Sciences of the United States of America 89 (14): 6428–32. July 1992. doi:10.1073/pnas.89.14.6428. PMID 1631139. Bibcode: 1992PNAS...89.6428O.
- ↑ "Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance". Science 264 (5157): 418–20. April 1994. doi:10.1126/science.8153629. PMID 8153629. Bibcode: 1994Sci...264..418H.
- ↑ "Solution structure of the DNA binding domain from Dead ringer, a sequence-specific AT-rich interaction domain (ARID)". The EMBO Journal 18 (21): 6084–94. November 1999. doi:10.1093/emboj/18.21.6084. PMID 10545119.
- ↑ "Solution structure of the ETS domain from murine Ets-1: a winged helix-turn-helix DNA binding motif". The EMBO Journal 15 (1): 125–34. January 1996. doi:10.2210/pdb1etc/pdb. PMID 8598195.
- ↑ "The ETS-domain transcription factor family". The International Journal of Biochemistry & Cell Biology 29 (12): 1371–87. December 1997. doi:10.1016/S1357-2725(97)00086-1. PMID 9570133.
- ↑ "The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution". Nature Structural Biology 8 (8): 710–4. August 2001. doi:10.1038/90429. PMID 11473263.
Further reading
- "Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins". Trends in Biochemical Sciences 14 (4): 137–40. April 1989. doi:10.1016/0968-0004(89)90145-X. PMID 2499084.
- "Winged helix proteins". Current Opinion in Structural Biology 10 (1): 110–6. February 2000. doi:10.1016/S0959-440X(99)00057-3. PMID 10679470.
- "A phylogenomic analysis of bacterial helix-turn-helix transcription factors". FEMS Microbiology Reviews 33 (2): 411–29. March 2009. doi:10.1111/j.1574-6976.2008.00154.x. PMID 19076237.
- "Chapter 1: Variation in form and function the helix-turn-helix regulators of the GntR superfamily". Advances in Applied Microbiology 69: 1–22. 2009. doi:10.1016/S0065-2164(09)69001-8. PMID 19729089.
- "The winged-helix DNA-binding motif: another helix-turn-helix takeoff". Cell 74 (5): 773–6. September 1993. doi:10.1016/0092-8674(93)90456-Z. PMID 8374950.
- "Prokaryotic transcription regulators: more than just the helix-turn-helix motif". Current Opinion in Structural Biology 12 (1): 98–106. February 2002. doi:10.1016/s0959-440x(02)00295-6. PMID 11839496.
External links
- Helix-turn-helix motif, lambda-like repressor, from EMBL
- Full PDB entry for PDB ID 1LMB
- Cro/C1-type HTH domain, more HTHs in PROSITE
Lua error: Internal error: The interpreter has terminated with signal "24".
| Bacterial regulatory helix-turn-helix protein, lysR family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_1 | ||||||||
| Pfam | PF00126 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR000847 | ||||||||
| PROSITE | PDOC00043 | ||||||||
| SCOP2 | 1al3 / SCOPe / SUPFAM | ||||||||
| |||||||||
| HTH DNA binding domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_10 | ||||||||
| Pfam | PF04967 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR007050 | ||||||||
| |||||||||
| Ribonuclease R winged-helix domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_12 | ||||||||
| Pfam | PF08461 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR013668 | ||||||||
| |||||||||
| HTH DNA binding domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_13 | ||||||||
| Pfam | PF11972 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR021068 | ||||||||
| |||||||||
| Helix-turn-helix | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_3 | ||||||||
| Pfam | PF01381 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR001387 | ||||||||
| PROSITE | PDOC50943 | ||||||||
| SCOP2 | 1r69 / SCOPe / SUPFAM | ||||||||
| |||||||||
| Bacterial regulatory protein, arsR family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_5 | ||||||||
| Pfam | PF01022 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR001845 | ||||||||
| PROSITE | PDOC00661 | ||||||||
| SCOP2 | 1smt / SCOPe / SUPFAM | ||||||||
| |||||||||
| Helix-turn-helix domain, rpiR family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_6 | ||||||||
| Pfam | PF01418 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR000281 | ||||||||
| PROSITE | PDOC51071 | ||||||||
| |||||||||
| Helix-turn-helix domain of resolvase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_7 | ||||||||
| Pfam | PF02796 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR006120 | ||||||||
| PROSITE | PDOC00334 | ||||||||
| SCOP2 | 1hcr / SCOPe / SUPFAM | ||||||||
| |||||||||
| Bacterial regulatory protein, Fis family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_8 | ||||||||
| Pfam | PF02954 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR002197 | ||||||||
| SCOP2 | 1f36 / SCOPe / SUPFAM | ||||||||
| |||||||||
| RNA polymerase III subunit RPC82 helix-turn-helix domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_9 | ||||||||
| Pfam | PF08221 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR013197 | ||||||||
| |||||||||
| Bacterial regulatory helix-turn-helix proteins, AraC family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_AraC | ||||||||
| Pfam | PF00165 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR000005 | ||||||||
| PROSITE | PDOC00040 | ||||||||
| SCOP2 | 1bl0 / SCOPe / SUPFAM | ||||||||
| |||||||||
| CodY helix-turn-helix domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_CodY | ||||||||
| Pfam | PF08222 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR013198 | ||||||||
| |||||||||
| DeoR-like helix-turn-helix domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_DeoR | ||||||||
| Pfam | PF08220 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR001034 | ||||||||
| PROSITE | PDOC00696 | ||||||||
| SCOP2 | 1lk5 / SCOPe / SUPFAM | ||||||||
| |||||||||
| IclR helix-turn-helix domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | HTH_IclR | ||||||||
| Pfam | PF09339 | ||||||||
| Pfam clan | CL0123 | ||||||||
| InterPro | IPR005471 | ||||||||
| PROSITE | PDOC00807 | ||||||||
| |||||||||
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".
Lua error: Internal error: The interpreter has terminated with signal "24".