Biology:Ganglion mother cell

Ganglion mother cells (GMCs) are cells involved in neurogenesis, in non-mammals, that divide only once to give rise to two neurons, or one neuron and one glial cell or two glial cells,[2] and are present only in the central nervous system. They are also responsible for transcription factor expression. While each ganglion mother cell necessarily gives rise to two neurons, a neuroblast can asymmetrically divide multiple times.[3] GMCs are the progeny of type I neuroblasts. Neuroblasts asymmetrically divide during embryogenesis to create GMCs.[4] GMCs are only present in certain species and only during the embryonic and larval stages of life. Recent research has shown that there is an intermediate stage between a GMC and two neurons. The GMC forms two ganglion cells which then develop into neurons or glial cells.[5] Embryonic neurogenesis has been extensively studied in Drosophila melanogaster embryos and larvae.
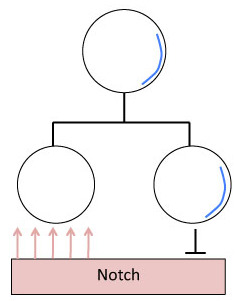
Mitotic division of Neuroblasts in Drosophila
The daughter cells of a neuroblast have two decidedly different neural fates. This is accomplished by neural fate determinants, important proteins that segregate asymmetrically. Most notable are Numb and Prospero. These proteins are evenly distributed in the neuroblast until mitosis occurs and they segregate totally into the newly formed GMC [6] During Mitosis Numb and Prospero localize to the basal cortex from which the GMC buds off.
- Numb is a suppressor of a signal protein called Notch. Suppressing Notch signaling allows the daughter cells to react to the same signal in different ways, allowing them to have different neural fates.
- Prospero is responsible for gene regulation in the GMC.
Both of these proteins co-function with adapter proteins that facilitate their transition to the basal cortex during Mitosis. These proteins are Miranda and Pon.
- Miranda localizes basally during interphase and then binds to Prospero, anchoring it to the basal cortex. Once the GMC has been created, Miranda releases Prospero, which evenly distributes throughout the new cell, and Miranda degrades.
- Pon also known as "partner of Numb" binds to Numb and co-localizes with it during Mitosis.
These four proteins act to inhibit self-renewal (the cell cycle) and promote differentiation (especially Prospero), which is why GMCs divide into their differentiated progeny instead of more GMCs.[3] Cell cycle progression is inhibited by Prospero because it activates cyclin-dependent kinase inhibitor (CKI).[5]
The vital differentiating proteins that are segregated into the daughter neuroblast and not the GMC are Bazooka, aPKC, Inscutable, and Partner of Inscutable (Pins). The proteins (with the exception of aPKC) form a ternary complex at the apical cortex independent of the proteins that segregate towards the basal cortex. The protein aPKC promotes self-renewal, encouraging the neuroblast to keep dividing and carry out its lineage.[3][6]
Research has suggested that certain tumor-suppressing proteins (Lgl, Dlg, or Brat) play a critical role in the asymmetric segregation of neural fate determinants and their localization to the basal cortex .[6] In clonal lines of neuroblasts that had been manipulated so that they lacked Lgl activity, Miranda did not segregate asymmetrically, but was evenly distributed throughout the cortex.
The temporal regulation of neuroblast asymmetric division is controlled by proteins Hunchback (Hb) and sevenup (svp). After division svp accumulates in both daughter cells and down-regulates Hb. In the GMC Prospero down-regulates svp, inhibiting the temporal trigger of cellular division.[7]
Type II Neuroblasts
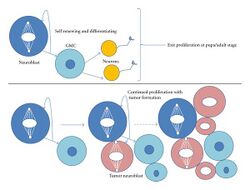
Type I neuroblasts have been more thoroughly observed and researched than type II. The main difference between them is that type II gives rise do a different kind of GMC (a Transit Amplifying GMC or TA-GMC, also known as intermediate progenitors), and its lineages are generally much longer.[3] TA-GMCs exhibits a different transcription factor from a generic GMC, Deadpan (Generic GMCs do in fact have Deadpan, but not outside of the nucleus). Type II neuroblasts do not contain detectable levels of Prospero. Unlike GMCs, TA-GMCs divide four to eight times, each time producing another TA-GMC and a generic GMC (which goes on to produce two neurons), which is why type II neuroblasts have a larger progeny than type I. Type II neuroblasts contribute a far larger population of neurons to the Drosophila brain.[1] Recent research has shown that type II lineages are more susceptible to tumor formation than type I. When experimentally knocking out proteins such as Numb or the tumor suppressing protein Brat the entire larval brain results in tumor formation only within type II lineages.[1] Tumor formation occurs when TA-GMCs revert to type II neuroblasts resulting in a highly increased cellular proliferation. The tumor phenotype can be suppressed with the introduction of ectopic Prospero. One of the main differences (perhaps the main difference) between type I and II neuroblasts is the presence of Prospero, suggesting that the introduction of Prospero can cause a type II neuroblast to transform into a type I identity.[1] It is also possible that Prospero simply inhibits the proliferation of type II neuroblasts without transforming them. Type I neuroblasts that have had the gene encoding for Prospero knocked out leads to tumor formation.[1]
Embryonic neural development in Drosophila
During the embryonic development of Drosophila, neuroblasts delaminate from their respective positions in the embryo and move towards the interior forming a ventral monolayer of cells, known as the neurogenic region.[4] The region is bilaterally symmetrical. The equivalent regions of neuronal growth in other common animal models do not have this symmetrical property, which makes Drosophila preferable for neurogenic study. The neurogenic region is composed of neuroblasts that divide and migrate throughout embryonic development. A larva embryo will contain about 30 neuroblasts per hemisegment of neurogenic tissue.[2] At a certain point, a neuroblast will undergo asymmetric cell division giving rise to a neuroblast and a ganglion mother cell. Each neuroblast can be traced through a lineage using methods such as green fluorescent protein transgene expression in order to investigate mechanisms of cellular diversity. A neuroblast lineage can produce as few as 3 GMCs or up to 20.[2] Research has been conducted to observe the movement of neuroblasts and GMCs in the neurogenic region during embryonic development using molecular markers.[4]
Specific Neuroblast lineages of interest
In Drosophila, each neural stem cell has been identified and categorized according to their location. Many neuroblasts, but not all, have also had their lineages identified (which GMCs they produce, and which subsequent neurons or glial cells the GMCs produce). For instance the first five GMCs of NB7-1 (the neuroblast located in the 7th row and first column of the cortex) sequentially generate the U1-U5 motor neurons, and then subsequently 30 interneurons. The first GMC of NB4-2 is known to produce motor neuron RP2.[8]
Post-embryonic neural development in Drosophila

The Drosophila CNS is composed of two brain hemispheres and the ventral ganglion.[5] Each hemisphere is composed of a laterally located Optic lobe (OL) and a medially located, generic Cerebrum (CB). At the end of embryonic development neuroblasts become quiescent, but re-enter their cell cycles during later specific larval stages.[5] The most complex structures in the insect/Drosophila brain, the central complex and the mushroom bodies, are responsible for associative learning and memory and do form during post-embryonic development.[10] Each OL is generated from three neuroepithelia called LPC (laminar precursor cells), OPC (outer proliferation center, and IPC (inner proliferation center). The OPC and IPC becomes asymmetric. Most of the development of the OL occurs at the end of the larval stage.[5] Prospero plays a different role in post embryonic neurogenesis than it did in the embryonic phase. Prospero is post-embryonically upregulated in order to promote neurons to exit the cell cycle, after GMCs differentiate during the embryogenesis Prospero is nearly undetectable.[5]
GMCs and mammalian neurogenic research
Mammalian neurogenic research has influenced further studies. Although there is no exact equivalent of GMCs in mammalian neurogenesis, mammalian neural stem cells do produce transit amplifying progenitors that expand neural population (similar to TA-GMCs).[3] An ortholog of Prospero in vertebrates (Prox1) is present in newly differentiating neurons and inhibits neural progenitor proliferation. This is similar to Prospero's effect type II neuroblasts that have expressed a tumor forming phenotype. The Prox1 protein is currently being studied as a candidate tumor suppression gene.[1]
Transcription factor expression
A common example of a transcription factor in neuroblasts is Deadpan, which promotes neural proliferation in the Optic lobe. A previously described transcription factor in GMCs is Prospero or Pros, a transcriptional repressor. It down-regulates cell cycle gene expression to restrict GMCs to one terminal mitosis. Pros is also present in young neurons, preventing mitotic action.[3] Prospero is not present in the progeny of GMCs and is thought to act as a timer, promoting prospective neurons out of their cell cycle.[5]
Implications
Studying neurogenesis in animal models such as Drosophila comes with many advantages and leads to a better understanding of relevant human neurogenic analogs such as neural stem cells. By obtaining a better understanding of how GMCs function and the role they play in neurogenesis, it may be possible to better understand their analogs in mammals.
See also
- Virtual fly brain
- Drosophila connectome
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Bayraktar, Boone, Drummond, Doe (2010). Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Development, 5:26.
- ↑ 2.0 2.1 2.2 Karcavich, Rachel, & Doe, Chris Q. (2005). Drosophila neuroblast 7-3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. The Journal of Comparative Neurology, 481(3), 240-251. Karcavich, Rachel E. (2005). Generating neuronal diversity in the Drosophila central nervous system: a view from the ganglion mother cells. Developmental Dynamics: An Official Publication Of The American Association Of Anatomists, 232(3), 609-616.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Doe, C. Q. et al (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. PMC 2804867.
- ↑ 4.0 4.1 4.2 Doe, C. Q. (1992). Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development, 116(4), 855-863.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 Colonques, Jordi, Ceron, Julian, Reichert, Heinrich, & Tejedor, Francisco J. (2011). A transient expression of Prospero promotes cell cycle exit of Drosophila postembryonic neurons through the regulation of Dacapo. PLoS ONE, 6(4), e19342-e19342.
- ↑ 6.0 6.1 6.2 Ohshiro, T., Yagami, T., Zhang, C., & Matsuzaki, F. (2000). Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature, 408(6812), 593-596.
- ↑ Mettler, Ulrike, Vogler, Georg, & Urban, Joachim. (2006). Timing of identity: spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero. Development, 133(3), 429-437.
- ↑ Grosskortenhaus, Robinson, Doe (2006). Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes and Development, 20(18): 2618–2627.
- ↑ Nidhi Saini and Heinrich Reichert, “Neural Stem Cells in Drosophila: Molecular Genetic Mechanisms Underlying Normal Neural Proliferation and Abnormal Brain Tumor Formation,” Stem Cells International, vol. 2012, Article ID 486169, 10 pages, 2012.
- ↑ Boyan, George, Williams, Leslie, Legl, Andrea, & Herbert, Zsofia. (2010). Proliferative cell types in embryonic lineages of the central complex of the grasshopper Schistocerca gregaria. Cell And Tissue Research, 341(2), 259-277.
 |