Biology:Gene transfer agent
Gene transfer agents (GTAs) are DNA-containing virus-like particles that are produced by some bacteria and archaea and mediate horizontal gene transfer. Different GTA types have originated independently from viruses in several bacterial and archaeal lineages. These cells produce GTA particles containing short segments of the DNA present in the cell. After the particles are released from the producer cell, they can attach to related cells and inject their DNA into the cytoplasm. The DNA can then become part of the recipient cells' genome.[1][2][3][4]
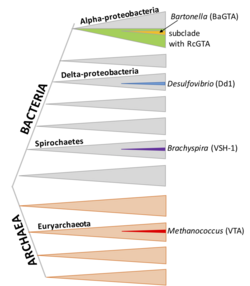
GTAs are classified as viriforms in the ICTV taxonomy. Among the GTAs mentioned by the article, RcGTA and DsGTA are now in the family Rhodogtaviriformidae, BaGTA in Bartogtaviriformidae, and VSH-1 in Brachygtaviriformidae.[5] Dd1 and VTA do not yet have a classification.
Discovery of gene transfer agents
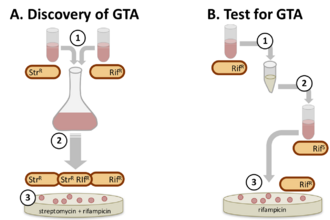
The first GTA system was discovered in 1974, when mixed cultures of Rhodobacter capsulatus strains produced a high frequency of cells with new combinations of genes.[6] The factor responsible was distinct from known gene-transfer mechanisms in being independent of cell contact, insensitive to deoxyribonuclease, and not associated with phage production. Because of its presumed function it was named gene transfer agent (GTA, now RcGTA) More recently other gene transfer agent systems have been discovered by incubating filtered (cell-free) culture medium with a genetically distinct strain.[3]
GTA genes and evolution
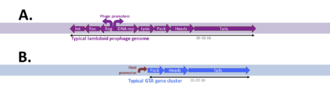
The genes specifying GTAs are derived from bacteriophage (phage) DNA that has integrated into a host chromosome. Such prophages often acquire mutations that make them defective and unable to produce phage particles. Many bacterial genomes contain one or more defective prophages that have undergone more-or less-extensive mutation and deletion. Gene transfer agents, like defective prophages, arise by mutation of prophages, but they retain functional genes for the head and tail components of the phage particle (structural genes) and the genes for DNA packaging. The phage genes specifying its regulation and DNA replication have typically been deleted, and expression of the cluster of structural genes is under the control of cellular regulatory systems. Additional genes that contribute to GTA production or uptake are usually present at other chromosome locations. Some of these have regulatory functions, and others contribute directly to GTA production (e.g. the phage-derived lysis genes) or uptake and recombination (e.g. production of cell-surface capsule and DNA transport proteins) These GTA-associated genes are often under coordinated regulation with the main GTA gene cluster.[7] Phage-derived cell-lysis proteins (holin and endolysin) then weaken the cell wall and membrane, allowing the cell to burst and release the GTA particles. The number of GTA particles produced by each cell is not known.
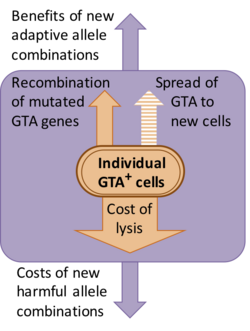
Some GTA systems appear to be recent additions to their host genomes, but others have been maintained for many millions of years. Where studies of sequence divergence have been done (dN/dS analysis), they indicate that the genes are being maintained by natural selection for protein function (i.e. defective versions are being eliminated).[8][9]
However the nature of this selection is not clear. Although the discoverers of GTA assumed that gene transfer was the function of the particles, the presumed benefits of gene transfer come at a substantial cost to the population. Most of this cost arises because GTA-producing cells must lyse (burst open) to release their GTA particles, but there are also genetic costs associated with making new combinations of genes, because most new combinations will usually be less fit than the original combination.[10] One alternative explanation is that GTA genes persist because GTAs are genetic parasites that spread infectiously to new cells. However this is ruled out because GTA particles typically are too small to contain the genes that encode them. For example, the main RcGTA cluster (see below) is 14 kb long, but RcGTA particles can contain only 4–5 kb of DNA.
Most bacteria have not been screened for the presence of GTAs, and many more GTA systems may await discovery. Although DNA-based surveys for GTA-related genes have found homologs in many genomes, but interpretation is hindered by the difficulty of distinguishing genes that encode GTAs from ordinary prophage genes.[8] [9]
GTA production
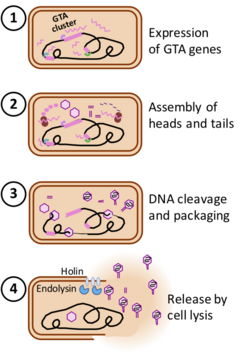
In laboratory cultures, production of GTAs is typically maximized by particular growth conditions that induce transcription of the GTA genes; most GTAs are not induced by the DNA-damaging treatments that induce many prophages. Even under maximally inducing conditions only a small fraction of the culture produces GTAs, typically less than 1%.[11][12]
The steps in GTA production are derived from those of phage infection. The structural genes are first transcribed and translated, and the proteins assembled into empty heads and unattached tails. The DNA packaging machinery then packs DNA into each head, cutting the DNA when the head is full, attaching a tail to the head, and then moving the newly-created DNA end on to a new empty head. Unlike prophage genes, the genes encoding GTAs are not excised from the genome and replicated for packaging in GTA particles. The two best studied GTAs (RcGTA and BaGTA) randomly package all of the DNA in the cell, with no overrepresentation of GTA-encoding genes.[11][13] The number of GTA particles produced by each cell is not known.
GTA-mediated transduction
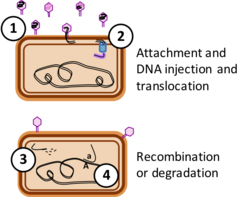
Whether release of GTA particles leads to transfer of DNA to new genomes depends on several factors. First, the particles must survive in the environment – little is known about this, although particles are reported to be quite unstable under laboratory conditions.[14] Second, particles must encounter and attach to suitable recipient cells, usually members of the same or a closely related species. Like phages, GTAs attach to specific protein or carbohydrate structures on the recipient cell surface before injecting their DNA. Unlike phage, the well-studied GTAs appear to inject their DNA only across the first of the two membranes surrounding the recipient cytoplasm, and they use a different system, competence-derived rather than phage-derived, to transport one strand of the double-stranded DNA across the inner membrane into the cytoplasm.[15][16]
If the cell's recombinational repair machinery finds a chromosomal sequence very similar to the incoming DNA, it replaces the former with the latter by homologous recombination, mediated by the cell's RecA protein. If the sequences are not identical this will produce a cell with a new genetic combination. However, if the incoming DNA is not closely related to DNA sequences in the cell it will be degraded, and the cell will reuse its nucleotides for DNA replication.
Specific GTA systems
RcGTA/Rhodobactegtaviriform (Rhodobacter capsulatus)
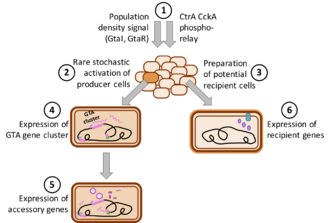
The GTA produced by the alphaproteobacterium Rhodobacter capsulatus, named R. capsulatus GTA (RcGTA), is currently the best studied GTA. When laboratory cultures of R. capsulatus enter stationary phase, a subset of the bacterial population induces production of RcGTA, and the particles are subsequently released from the cells through cell lysis.[12] Most of the RcGTA structural genes are encoded in a ~ 15 kb genetic cluster on the bacterial chromosome. However, other genes required for RcGTA function, such as the genes required for cell lysis, are located separately.[2][17] RcGTA particles contain 4.5 kb DNA fragments, with even representation of the whole chromosome except for a 2-fold dip at the site of the RcGTA gene cluster.
Regulation of GTA production and transduction has been best studied in R. capsulatus, where a quorum-sensing system and a CtrA-phosphorelay control expression of not only the main RcGTA gene cluster, but also a holin/endolysin cell lysis system, particle head spikes, an attachment protein (possibly tail fibers), and the capsule and DNA processing genes needed for RcGTA recipient function. An uncharacterized stochastic process further limits expression of the gene cluster is to only 0.1-3% of the cells.
RcGTA-like clusters are found in a large subclade of the alphaproteobacteria, although the genes also appear to be frequently lost by deletion. Recently, several members of the order Rhodobacterales have been demonstrated to produce functional RcGTA-like particles. Groups of genes with homology to the RcGTA are present in the chromosomes of various types of alphaproteobacteria.[8]
DsGTA/Dinogtaviriform (Dinoroseobacter shibae)
D. shibae, like R. capsulatus, is a member of the Order Rhodobacterales, and its GTA shares a common ancestor and many features with RcGTA, including gene organization, packaging of short DNA fragments (4.2 kb) and regulation by quorum sensing and a CtrA phosphorelay.[18] However, its DNA packaging machinery has much more specificity, with sharp peaks and valleys of coverage suggestion that it may preferentially initiate packaging at specific sites in the genome. The DNA of the major DsGTA gene cluster is packaged very poorly.
BaGTA/Bartonegtaviriform (Bartonella species)
Bartonella species are members of the Alphaproteobacteria like R. capsulatus and D. shibae, but BaGTA is not related to RcGTA and DsGTA.[19] BaGTA particles are larger than RcGTA and contain 14 kb DNA fragments. Although this capacity could in principle allow BaGTA to package and transmit its 14 kb GTA cluster, measurements of DNA coverage show reduced coverage of the cluster. An adjacent region of high coverage is thought to be due to local DNA replication.[13]
VSH-1 (Brachyspira hyodysenteriae)
Brachyspira is a genus of spirochete; several species have been shown to carry homologous GTA gene clusters. Particles contain 7.5 kb DNA fragments. Production of VSH-1 is stimulated by the DNA-damaging agent mitomycin C and by some antibiotics. It is also associated with detectable cell lysis, indicating that a substantial fraction of the culture may be producing VSH-1.[20]
Dd1 (Desulfovibriondesulfuricans)
D. desulfuricans is a soil bacterium in the deltaproteobacteria; Dd1 packages 13.6 kb of DNA fragments. It is unclear which genes encode for this GTA: there is one 17.8 kb area with phage-like structual genes in the bacterial genome, but their link to GTA production is not yet experimentally proven.[21]
VTA (Methanococcus voltae)
M. voltae is an archaean; its GTA is known to transfer 4.4 kb DNA fragments but has not been otherwise characterized,[22] although a defective provirus related to Methanococcus head-tailed viruses (Caudoviricetes) in M. voltae A3 genome has been suggested to represent the GTA locus.[23] A possible terL terminase (D7DSG2) was again identified in 2019.[24]
See also
- Gene transfer agent-release holin family
- Horizontal gene transfer
References
- ↑ "The Distribution, Evolution, and Roles of Gene Transfer Agents in Prokaryotic Genetic Exchange". Annual Review of Virology 4 (1): 87–104. September 2017. doi:10.1146/annurev-virology-101416-041624. PMID 28784044.
- ↑ 2.0 2.1 "Gene transfer agents: phage-like elements of genetic exchange". Nature Reviews. Microbiology 10 (7): 472–82. June 2012. doi:10.1038/nrmicro2802. PMID 22683880.
- ↑ 3.0 3.1 "Prophage-like gene transfer agents-novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species". Anaerobe 13 (2): 43–9. April 2007. doi:10.1016/j.anaerobe.2007.03.004. PMID 17513139.
- ↑ "Small extracellular particles with big potential for horizontal gene transfer: membrane vesicles and gene transfer agents". FEMS Microbiology Letters 365 (19). October 2018. doi:10.1093/femsle/fny192. PMID 30085064.
- ↑ Kogay, R; Koppenhöfer, S; Beatty, JT; Kuhn, JH; Lang, AS; Zhaxybayeva, O (2022). "Formal recognition and classification of gene transfer agents as viriforms.". Virus Evolution 8 (2): veac100. doi:10.1093/ve/veac100. PMID 36381234.
- ↑ "Genetic recombination in Rhodopseudomonas capsulata". Proceedings of the National Academy of Sciences of the United States of America 71 (3): 971–3. March 1974. doi:10.1073/pnas.71.3.971. PMID 4522805. Bibcode: 1974PNAS...71..971M.
- ↑ "Guaranteeing a captive audience: coordinated regulation of gene transfer agent (GTA) production and recipient capability by cellular regulators". Current Opinion in Microbiology 38: 122–129. August 2017. doi:10.1016/j.mib.2017.05.003. PMID 28599143.
- ↑ 8.0 8.1 8.2 "Insights into origin and evolution of α-proteobacterial gene transfer agents". Virus Evolution 3 (2): vex036. July 2017. doi:10.1093/ve/vex036. PMID 29250433.
- ↑ 9.0 9.1 "Origin and Evolution of the Bartonella Gene Transfer Agent". Molecular Biology and Evolution 35 (2): 451–464. February 2018. doi:10.1093/molbev/msx299. PMID 29161442.
- ↑ "Evolution of Bacterial Gene Transfer Agents" (in en). Frontiers in Microbiology 9: 2527. 2018. doi:10.3389/fmicb.2018.02527. PMID 30410473.
- ↑ 11.0 11.1 "DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA". Molecular Microbiology 85 (2): 314–25. July 2012. doi:10.1111/j.1365-2958.2012.08113.x. PMID 22640804.
- ↑ 12.0 12.1 Banfield, Bruce W, ed (2012). "One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus". PLOS ONE 7 (8): e43772. doi:10.1371/journal.pone.0043772. PMID 22916305. Bibcode: 2012PLoSO...743772F.
- ↑ 13.0 13.1 "Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii". PLOS Genetics 5 (7): e1000546. July 2009. doi:10.1371/journal.pgen.1000546. PMID 19578403.
- ↑ Marrs, B.; Yen, H. C.; Solioz, M. (1975-08-01). "Release and uptake of gene transfer agent by Rhodopseudomonas capsulata." (in en). Journal of Bacteriology 123 (2): 651–657. doi:10.1128/jb.123.2.651-657.1975. ISSN 1098-5530. PMID 1150627.
- ↑ "Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA)". Molecular Microbiology 87 (4): 802–17. February 2013. doi:10.1111/mmi.12132. PMID 23279213.
- ↑ "Homologues of Genetic Transformation DNA Import Genes Are Required for Rhodobacter capsulatus Gene Transfer Agent Recipient Capability Regulated by the Response Regulator CtrA". Journal of Bacteriology 197 (16): 2653–63. August 2015. doi:10.1128/JB.00332-15. PMID 26031909.
- ↑ "Phosphate concentration and the putative sensor kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent". Journal of Bacteriology 195 (22): 5025–40. November 2013. doi:10.1128/JB.00669-13. PMID 23995641.
- ↑ "Packaging of Dinoroseobacter shibae DNA into Gene Transfer Agent Particles Is Not Random". Genome Biology and Evolution 10 (1): 359–369. January 2018. doi:10.1093/gbe/evy005. PMID 29325123.
- ↑ "Gene Transfer Agent Promotes Evolvability within the Fittest Subpopulation of a Bacterial Pathogen". Cell Systems 4 (6): 611–621.e6. June 2017. doi:10.1016/j.cels.2017.05.011. PMID 28624614.
- ↑ "Identification of genes associated with prophage-like gene transfer agents in the pathogenic intestinal spirochaetes Brachyspira hyodysenteriae, Brachyspira pilosicoli and Brachyspira intermedia". Veterinary Microbiology 134 (3–4): 340–5. March 2009. doi:10.1016/j.vetmic.2008.09.051. PMID 18950961. https://researchrepository.murdoch.edu.au/id/eprint/7883/.
- ↑ Lang, AS; Zhaxybayeva, O; Beatty, JT (11 June 2012). "Gene transfer agents: phage-like elements of genetic exchange.". Nature Reviews. Microbiology 10 (7): 472–82. doi:10.1038/nrmicro2802. PMID 22683880.
- ↑ "Transduction-like gene transfer in the methanogen Methanococcus voltae". Journal of Bacteriology 181 (10): 2992–3002. May 1999. doi:10.1128/JB.181.10.2992-3002.1999. PMID 10321998.
- ↑ Krupovic, M; Forterre, P; Bamford, DH (2010). "Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria.". Journal of Molecular Biology 397 (1): 144–60. doi:10.1016/j.jmb.2010.01.037. PMID 20109464.
- ↑ Sherlock, D; Leong, JX; Fogg, PCM (1 December 2019). "Identification of the First Gene Transfer Agent (GTA) Small Terminase in Rhodobacter capsulatus and Its Role in GTA Production and Packaging of DNA.". Journal of Virology 93 (23). doi:10.1128/JVI.01328-19. PMID 31534034.
 |
