Biology:HIKESHI
 Generic protein structure example |
HIKESHI is a protein important in lung and multicellular organismal development[1] that, in humans, is encoded by the HIKESHI gene.[2] HIKESHI is found on chromosome 11 in humans and chromosome 7 in mice. Similar sequences (orthologs) are found in most animal and fungal species. The mouse homolog, lethal gene on chromosome 7 Rinchik 6 protein is encoded by the l7Rn6 gene.[3]
Gene
HIKESHI is a protein-coding gene in Homo sapiens. Alternate names for the gene are FLJ43020, HSPC138, HSPC179, and L7RN6. Located on long arm of chromosome 11 at area q14.2, the entire gene including introns and exons is 42,698 base pairs on the plus strand. The mRNA of HIKESHI Variant 1 includes exons 1, 3, 4, 5, and 7 amounting to 1,183 base pairs, with base pairs 239 to 832 representing the coding regions.
Alternative Splicing
Variant 1 is the longest and most common protein coding variant. The three other main variants use an alternate exon sequence that throws off the reading frame, causing early termination of the mRNA sequence and undergoes protein decay. The table below shows the different variants and exon usage.
| Variant | Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | Protein Coding |
|---|---|---|---|---|---|---|---|---|
| 1 | x | x | x | x | x | Yes | ||
| 2 | x | x | x | x | x | x | No | |
| 3 | x | x | x | x | x | No | ||
| 4 | x | x | x | x | No |
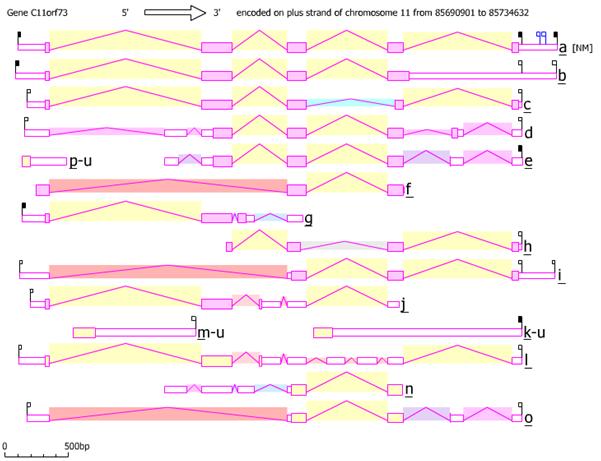
The four variants shown in the table above are the most common isoforms found in human cells. There are a total of 13 alternatively spliced sequences and three unspliced forms that utilize two alternative promoters. The mRNA variants differ on the combination of 8 different exons, alternate, overlapping exons, and the retention of introns. Besides alternative splicing, the mRNAs differ by truncation on the 3’ end. Variant 1 is one of ten mRNAs that has been shown to code for a protein, while the rest seem bound for nonsense mediated mRNA decay.
AceView[4] representation of C11orf73 isoforms
Promoter
The Promoter region, GXP 47146, was found using the ElDorado[5] tool from Genomatix. The 840 bp sequence is located before the HIKESHI gene at DNA points 86012753 to 86013592. The promoter is conserved in 12 of 12 orthologs and codes for 6 relevant transcripts.
Conserved transcription factor binding sites from Genomatix ElDorado tool:
| Detailed Family Information | From | To | Anchor | Orientation | Conserved in Mus Musculus | Matrix Sim | Sequence | Occurrence |
|---|---|---|---|---|---|---|---|---|
| Cell cycle regulators: Cell cycle homology element | 137 | 149 | 143 | + strand | conserved | 0.943 | ggacTTGAattca | 1 |
| GATA binding factors | 172 | 184 | 178 | + strand | conserved | 0.946 | taaAGATttgagg | 1 |
| Vertebrate TATA binding protein factor | 193 | 209 | 201 | + strand | conserved | 0.983 | tcctaTAAAatttggat | 1 |
| Heat schock factors | 291 | 315 | 303 | + strand | conserved | 0.992 | cacagaaacgttAGAAgcatctctt | 4 |
| Human and murine ETS1 factors | 512 | 532 | 522 | + strand | conserved | 0.984 | taagccccGGAAgtacttgtt | 3 |
| Zinc finger transcription factor RU49, Zipro1 | 522 | 528 | 525 | + strand | conserved | 0.989 | aAGTAct | 2 |
| Krueppel like transcription factors | 618 | 634 | 626 | + strand | conserved | 0.925 | tggaGGGGcagacaccc | 1 |
| SOX/SRY-sex/testis determining and HMG box factors | 636 | 658 | 647 | + strand | conserved | 0.925 | cccgcaAATTctggaaggttctt | 1 |
Termination
Termination of the mRNA product is encoded for within the cDNA of the gene. The end termination of an mRNA product generally has three main features: the poly A signal, the poly A tail, and an area of sequence that can form a stem loop structure. The poly A signal is a highly conserved site, six nucleotide long sequence. In eukaryotes the sequence is AATAAA and is located about 10–30 nucleotides from the poly A site. The AATAAA sequence is a highly conserved, eukaryotic polyA signal that signals for polyadenylation of the mRNA product 10–30 base pairs after the signal sequence. The polyA site for C11orf73 is GTA.
Gene expression
HIKESHI was determined to be expressed ubiquitously at a high level of 2.3 times above the average. C11orf73 is expressed in a large number of human tissues.[6][7] Between the Expression Profiles and the EST Profile on UniGene, only 11 tissues were shown not to express C11orf73, most likely due to small sample sizes in the tissue.
Protein
The human HIKESHI gene encodes for a protein called uncharacterized protein C11orf73.[2] The homologous mouse L7rn6 gene encodes a protein called lethal gene on chromosome 7 Rinchik 6.[3]
1 mfgclvagrl vqtaaqqvae dkfvfdlpdy esinhvvvfm lgtipfpegm ggsvyfsypd 61 sngmpvwqll gfvtngkpsa ifkisglksg egsqhpfgam nivrtpsvaq igisvellds 121 maqqtpvgna avssvdsftq ftqkmldnfy nfassfavsq aqmtpspsem fipanvvlkw 181 yenfqrrlaq nplfwkt
The encoded human protein is 197 amino acids long and weighs 21,628 daltons. Through analogy to the mouse protein, the hypothetical function of the human HIKESHI protein is the organization and function of the secretory apparatus in lung cells.[1]
| Protein of unknown function (DUF775) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | DUF775 | ||||||||
| Pfam | PF05603 | ||||||||
| InterPro | IPR008493 | ||||||||
| |||||||||
The protein domain known as DUF775 (Domain of Unknown Function 775) is located within both the human HIKESHI and mouse L7rn6 proteins. The DUF775 domain is 197 amino acids long, the same length as the protein. Other proteins that make up the DUF 775 super family by definition include all the orthologs of C11orf73.
Hydropathy analysis shows that there are no extensive hydrophobic regions in the protein and, hence, it is concluded that HIKESHI is a cytoplasmic protein. The isoelectric point for C11orf73 is 5.108 suggesting it functions optimally in a more acidic environment.
SNP
The only SNP,[9] or single-nucleotide polymorphism, for the C11orf73 sequence results in an amino acid change within the protein. The lack of other SNPs are most likely due to the high level of conservation of HIKESHI and the lethal effect a mutation in the protein bestows upon the organism. The phenotype for the SNP is unknown.
| Function | dbSNP Allele | Protein Residue | Codon Position | Amino Acid Position |
|---|---|---|---|---|
| Reference | C | Proline [P] | 1 | 47 |
| Missense | G | Alanine [A] | 1 | 47 |
Gene Neighborhood
The surrounding genes of HIKESHI are CCDC81, ME3, and EED. The genetic neighborhood is looked at in order to get a better understanding of the possible function of the gene by looking at the function of the surrounding genes.
The CCDC81 gene codes for an uncharacterized protein product and is oriented on the plus strand. CCDC81stands for coiled-coil domain containing 81 isoform 1.
The ME3 gene stands for mitochondrial malic enzyme 3 precursor. Malic enzyme catalyzes the oxidative decarboxylation of malate to pyruvate using either NAD+ or NADP+ as a cofactor. Mammalian tissues contain 3 distinct isoforms of malic enzyme: a cytosolic NADP(+)-dependent isoform, a mitochondrial NADP(+)-dependent isoform, and a mitochondrial NAD(+)-dependent isoform. This gene encodes a mitochondrial NADP(+)-dependent isoform. Multiple alternatively spliced transcript variants have been found for this gene, but the biological validity of some variants has not been determined.[11]
The EED gene stands for embryonic ectoderm development isoform b and is a member of the Polycomb-group (PcG) family. PcG family members form multimeric protein complexes, which are involved in maintaining the transcriptional repressive state of genes over successive cell generations. This protein interacts with enhancer of zeste 2, the cytoplasmic tail of integrin beta7, immunodeficiency virus type 1 (HIV-1) MA protein, and histone deacetylase proteins. This protein mediates repression of gene activity through histone deacetylation, and may act as a specific regulator of integrin function. Two transcript variants encoding distinct isoforms have been identified for this gene.[12]
Interactions
The programs STRING[13] and Sigma-Aldrich's Favorite Gene[14] suggested possible protein interactions with C11orf73. ARGUL1, CRHBP, and EED were derived from textmining and HNF4A came from Sigma-Aldrich.
| Protein | Description | Method | Score |
|---|---|---|---|
| ARGUL1 | Unknown | Textmining | 0.712 |
| CRHBP | Corticotropin releasing hormone binding protein | Textmining | 0.653 |
| EED | Embryonic ectoderm development | Textmining | 0.420 |
| HNF4A | Transcription regulator | Sigma-Aldrich | N/A |
ARGUL1 is an unknown protein with an unknown function. CRHBP is a corticotrophin releasing hormone binding protein which could possibly play a role in a signal cascade that involves or activates HIKESHI. EED, a neighboring protein of C11orf73, is an embryonic ectoderm development protein and is a member of the Polycomb-group (PcG) family. PcG family members form multimeric protein complexes, which are involved in maintaining the transcriptional repressive state of genes over successive cell generations. HNF4A is a transcription regulator and it is unknown if HNF4A regulates C11orf73's expression or simply interacts with it.[12
Evolutionary History
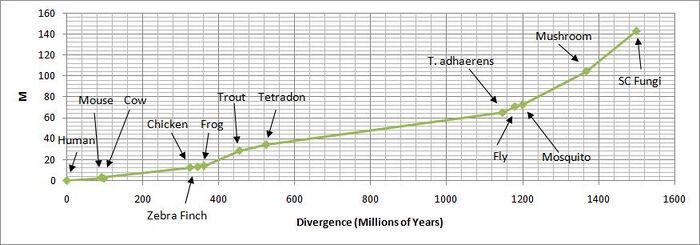
The evolutionary history of organisms can be determined using the sequences of orthologs as time references to create a phylogenetic tree. The CLUSTALW[15] compares multiple sequences, the program can also be used to create such a phylogenetic tree based on the orthologs of C11orf73. The tree to the right shows the generated phylogenetic tree with a time line based on time of divergence. The tree made from the HIKESHI orthologs is identical to the literature phylogenetic tree, even grouping together similar organisms such as fish, birds, and fungi.
Orthologs
Homologous sequences are orthologous if they were separated by a speciation event: when a species diverges into two separate species, the divergent copies of a single gene in the resulting species are said to be orthologous. Orthologs, or orthologous genes, are genes in different species that are similar to each other because they originated from a common ancestor. Orthologous sequences provide useful information in taxonomic classification and phylogenetic studies of organisms. The pattern of genetic divergence can be used to trace the relatedness of organisms. Two organisms that are very closely related are likely to display very similar DNA sequences between two orthologs. Conversely, an organism that is further removed evolutionarily from another organism is likely to display a greater divergence in the sequence of the orthologs being studied.
Table of Chromosome 11 open reading frame 73 Orthologs
| Species | Common Name | Protein Name | Accession Number | NT Length | NT Identity | AA Length | AA Identity | E-Value |
|---|---|---|---|---|---|---|---|---|
| Homo sapiens | Human | C11orf73 | NM_016401 | 1187 bp | 100% | 197 aa | 100% | 0 |
| Bon taurus | Cow | LOC504867 | NP_001029398 | 996 bp | 73.60% | 197 aa | 98% | 5.30E-84 |
| Mus musculus | Mouse | l7Rn6 | NP_080580 | 1045 bp | 72.90% | 197 aa | 97% | 4.80E-83 |
| Gallus gallus | Chicken | LOC427034 | N/A | 851 bp | 56.20% | 197 aa | 88.3% | 5.60E-76 |
| Taeniopygia guttata | Zebra Finch | LOC100190155 | ACH44077 | 997 bp | 61.60% | 997 aa | 87.80% | 1.20E-75 |
| Xenopus laevis | Frog | MGC80709 | NP_001087012 | 2037 bp | 36.50% | 197 aa | 86.80% | 1.70E-75 |
| Oncorhynchus mykiss | Rainbow Trout | CK073 | NP_001158574 | 940 bp | 52.20% | 197 aa | 75.10% | 2.70E-66 |
| Tetradon nigroviridis | Tetradon | unnamed protein product | CAF89643 | N/A | N/A | 197 aa | 70.90% | 1.40E-61 |
| Trichoplax adhaerens | Trichoplax adhaerens | TRIADDRAFT_19969 | XP_002108733 | 600 bp | 33.10% | 199 aa | 52.30% | 2.00E-47 |
| Culex quinquefasciatus | Mosquito | conserved hypothetical protein | XP_001843282 | 594 bp | 30.70% | 197 aa | 49.30% | 2.50E-41 |
| Drosophilia melanogaster | Fly | CG13926 | NP_647633 | 594 bp | 31.50% | 197 aa | 48.50% | 4.50E-39 |
| Laccaria bicolor | Mushroom | predicted protein | XP_001878996 | 696 bp | 36.40% | 202 aa | 35.20% | 8.30E-24 |
| Candida albicans | Fungi | CaO19.13758 | XP_716157 | 666 bp | 36.10% | 221 aa | 24% | 5.70E-11 |
The table shows the 13 sequences (12 orthologs, 1 original sequence) along with protein name, accession numbers, nucleotide identity, protein identity, and E-values. The accession numbers are the identification numbers from the NCBI Protein database. The nucleotide sequence can be accessed from the protein's sequence page from DBSOURCE, which gives the accession number and is a link to the nucleotide's sequence page. The length of both the nucleotide and protein sequence for each ortholog and its respective organism are listed in the table as well. Next to the sequence lengths are the identities of the ortholog to the original HIKESHI gene. The identities and E-values were acquired using the global alignment program, ALIGN, from the SDSC Biology Workbench and BLAST from NCBI.
The graph shows the percent identity of the ortholog against the divergence time of the organism to produce a mostly linear curve. The two main joints within the curve suggest times of gene duplication, around 450 million years and 1150 million years ago respectively. The paralogs from the gene duplications are probably so dissimilar from the highly conserved orthologs of HIKESHI that it was not found using the Blink or BLAST tools.
The value m (total number of amino acid changes that have occurred in a 100 amino acid segment), which is the corrected value of n (number of amino acid differences from the template sequence), is also used to calculate λ (the average amino acid changes per year, usually represented in values of λE9).
m/100 = –ln(1-n/100) λ = (m/100)/(2*T)
References
- ↑ 1.0 1.1 "l7Rn6 Encodes a Novel Protein Required for Clara Cell Function in Mouse Lung Development". Genetics 172 (1): 389–99. January 2006. doi:10.1534/genetics.105.048736. PMID 16157679.
- ↑ 2.0 2.1 "Cloning and Functional Analysis of cDNAs with Open Reading Frames for 300 Previously Undefined Genes Expressed in CD34+ Hematopoietic Stem/Progenitor Cells". Genome Res. 10 (10): 1546–60. October 2000. doi:10.1101/gr.140200. PMID 11042152.
- ↑ 3.0 3.1 "N-ethyl-N-nitrosourea-induced prenatally lethal mutations define at least two complementation groups within the embryonic ectoderm development (eed) locus in mouse chromosome 7". Mamm. Genome 4 (7): 349–53. 1993. doi:10.1007/BF00360583. PMID 8358168. https://zenodo.org/record/1232417.
- ↑ AceView NCBI Gene Information AceView
- ↑ Genomatix ElDorade tool for promoter analysis ElDorado Product Page
- ↑ "Expression profile for C11orf73". GeneNote version 2.4. Weizmann Institute of Science. September 2009. http://bioinfo2.weizmann.ac.il/cgi-bin/genenote/GN_results.pl?keyword_type=2_gc_id&data_type=norm2&results=yes&keyword=GC11P085691.
- ↑ "EST Profile - Hs.283322". National Center for Biotechnology Information, United States National Library of Medicine. https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.283322.
- ↑ Saier Lab Bioinformatics Group http://www.tcdb.org/progs/hydro.php
- ↑ NCBI SNP Database https://www.ncbi.nlm.nih.gov/snp/
- ↑ NCBI Entrez https://www.ncbi.nlm.nih.gov/nuccore/NC_000011.9?from=86011067&to=86059171&report=graph
- ↑ RefSeq NCBI Database https://www.ncbi.nlm.nih.gov/RefSeq/
- ↑ "RefSeq: NCBI Reference Sequence Database". https://www.ncbi.nlm.nih.gov/RefSeq/.
- ↑ STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) http://string-db.org/
- ↑ Sigma-Aldrich's Favorite Gene http://www.sigmaaldrich.com/life-science/your-favorite-gene-search.html
- ↑ CLUSTALW Program Julie D. Thompson, Desmond G. Higgins and Toby J. Gibson http://workbench.sdsc.edu/
External links
- Human C11orf73 genome location and C11orf73 gene details page in the UCSC Genome Browser.
 |