Biology:Haplogroup E-V12
| Haplogroup E-V12 | |
|---|---|
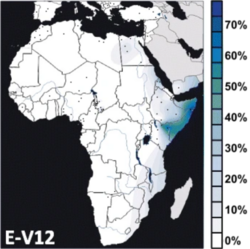 Interpolated frequency distribution.[1] | |
| Possible time of origin | c. 12,300 years BP[2] |
| Coalescence age | c. 10,500 years BP[2] |
| Possible place of origin | Lower Egypt/Libya[3] or Upper Egypt/Northern Sudan[4] |
| Ancestor | E-V68/M78[5] |
| Defining mutations | V12, Z1216[5] |
The human Y-chromosome haplogroup E-V12 is a subclade of E-M78, which in turn is part of the larger haplogroup E1b1b1.[3] According to Cruciani et al. (2007), the E-V12 sublineage likely originated in Northern Africa. It has two main branches: E-V32 which is most common in the Horn of Africa, and E-CTS693 (previously known as E-V12*) which is most common in Upper Egypt and to a lesser extent in Sudan. E-CTS693 is also scattered in low frequencies across the Levant, Anatolia, the Central Sahel, the African Great Lakes region, and Europe.[6][3]
Ancient DNA
- One individual from Bronze Age Alalakh dated c. 1878-1637 BC belonged to haplogroup E-V12.[7] Alalakh was a city in northern Levant settled by Amorites and some Hurrians.[7]
- One individual from imperial Rome dated c. 2000-1800 ybp (years before present) belonged to haplogroup E-V12.[8]
- A 1st century Nubian mercenary genome from Serbia (Roman Empire) carried haplogroup E-V32 and L2a1j. On the PCA, the outlier individual clustered with present day East African populations.[9]
- One Medieval individual dated c. 1130-520 ybp from Villa Magna, Central Italy belonged to haplogroup E-V12.[8]
Undifferentiated E-V12* lineages
Undifferentiated E-V12* lineages (not E-V32 or E-M224, so therefore named "E-V12*") peak in frequency among Southern Egyptians (up to 74.5%).[10] The subclades are also scattered widely in small amounts in both Northern Africa and Europe, but with very little sign in Western Asia, apart from Turkey.[3] These E-V12* lineages were formerly included (along with many E-V22* lineages[Note 1]) in Cruciani et al.'s original (2004) "delta cluster", which he had defined using Y-STR profiles. With the discovery of the defining SNP, Cruciani et al. (2007) reported that V12* was found in its highest concentrations in Egypt, especially Southern Egypt.
Hassan et al. (2008) report a significant presence of E-V12* in neighboring Sudan, including 5/33 Copts and 5/39 Nubians. E-V12* made up approximately 20% of the Sudanese E-M78. They propose that the E-V12 and E-V22 sub-clades of E-M78 might have been brought to Sudan from their place of origin in North Africa after the progressive desertification of the Sahara around 6,000–8,000 years ago. Sudden climate change might have forced several Neolithic cultures/people to migrate northward to the Mediterranean and southward to the Sahel and the Nile Valley.[11] The E-V12* paragroup is also observed in Europe (e.g. amongst French Basques) and Eastern Anatolia (e.g. Erzurum Turks).[3]
The non-basal subhaplogroup E1b1b-V12/E3b1a1 has been found at highest frequencies among various Afroasiatic-speaking populations in eastern Africa, including Garreh (74.1%), Gabra (58.6%), Wata (55.6%), Borana (50.0%), Sanye (41.7%), Beja (33.3%) and Rendille (29.0%).[12]
Sub-clades of E-V12
E-V32
Cruciani et al. (2007) suggest that this sub-clade of E-V12, which originated in North Africa, and then subsequently expanded further south into the Horn of Africa, where it is now prevalent,[Note 2] with speakers of Cushitic. Before the discovery of V32, Cruciani et al. (2004) referred to the same lineages as the "gamma cluster", which was estimated to have arisen about 8,500 years ago. They stated that "the highest frequencies in the three Cushitic-speaking groups: the Borana from Kenya (71.4%), the Oromo from Ethiopia (32.0%), and the Somali (80%). Outside of eastern Africa, it was found only in two subjects from Egypt (3.6%) and in one Arab from Morocco". Sanchez et al. (2005) found it extremely prominent in Somali men and stated that "the male Somali population is a branch of the Horn African population – closely related to the Oromos in Ethiopia and North Kenya (Boranas)" and that their gamma cluster lineages "probably were introduced into the Somali population 4000–5000 years ago". More recently, Tillmar et al. (2009) typed 147 males from Somalia for 12 Y-STR loci, and observed that 77% (113/147) had typical E-V32 haplotypes. This is currently the highest frequency of E-V32 found in any single sample population. Similarly, Hassan et al. (2008) in their study observed this to be the most common of the sub-clades of E-M78 found in Sudan, especially among the Beja, Masalit and Fur. The Beja, like Somalis and Oromos, speak an Afro-Asiatic language and live along the "corridor" from the Horn of Africa to Egypt. Hassan et al. (2008) interpret this as reinforcing the "strong correlation between linguistic and genetic diversity" and signs of relatedness between the Beja and the peoples of the Horn of Africa such as the Amhara, Oromo, and Somalis. On the other hand, the Masalit and Fur live in Darfur and speak a Nilo-Saharan language. The authors observed in their study that "the Masalit possesses by far the highest frequency of the E-M78 and of the E-V32 haplogroup", which they believe suggests "either a recent bottleneck in the population or a proximity to the origin of the haplogroup." However, More recently, Tillmar et al. (2009) typed 147 males from Somalia for 12 Y-STR loci, and observed that 77% (113/147) had typical E-V32 haplotypes. This is the highest frequency of E-V32 found in any single sample population.
The STR data from Cruciani et al. (2007) concerning E-V12 can be summarized as follows.
| Haplotype | description | YCAIIa | YCAIIb | DYS413a | DYS413b | DYS19 | DYS391 | DYS393 | DYS439 | DYS460 | DYS461 | A10 |
| E-V12* | modal | 19 | 22 | 22 | 22 | 13 | 10 | 13 | 11 | 11 | 9 | 13 |
| min | 18 | 21 | 20 | 21 | 11 | 10 | 12 | 11 | 8 | 8 | 11 | |
| max | 19 | 22 | 22 | 23 | 15 | 12 | 14 | 13 | 12 | 10 | 14 | |
| number | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | 40 | |
| E-V32 | modal | 19 | 21 | 22 | 23 | 11 | 10 | 13 | 12 | 10 | 10 | 13 |
| min | 19 | 19 | 20 | 21 | 11 | 9 | 12 | 11 | 9 | 10 | 11 | |
| max | 20 | 22 | 22 | 24 | 11 | 11 | 13 | 13 | 12 | 11 | 14 | |
| number | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | 35 | |
| All E-V12 | modal | 19 | 22 | 22 | 23 | 11 | 10 | 13 | 11 | 11 | 10 | 13 |
| min | 18 | 19 | 20 | 21 | 11 | 9 | 12 | 11 | 8 | 8 | 11 | |
| max | 20 | 22 | 22 | 24 | 15 | 12 | 14 | 13 | 12 | 11 | 14 | |
| number | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | |
Sub-clades
Notes
- ↑ Cruciani et al. (2004): "E-V22 and E-V12* chromosomes are intermingled and not clearly differentiated by their microsatellite haplotypes". In Cruciani et al. (2007) the same authors show that a branch of E-V13 found amongst the Druze Arabs is also in the delta cluster. (Contrast the data tables of Cruciani et al. (2007) and Cruciani et al. (2004).)
- ↑ Cruciani et al. (2007): Fig. 2/C
References
- ↑ D'Atanasio E et al. (2018). "The peopling of the last Green Sahara revealed by high-coverage resequencing of trans-Saharan patrilineages.". Genome Biol 19 (1): 20. doi:10.1186/s13059-018-1393-5. PMID 29433568.
- ↑ 2.0 2.1 "E-V12 YTree". https://www.yfull.com/tree/E-V12.
- ↑ 3.0 3.1 3.2 3.3 3.4 Cruciani et al. (2007)
- ↑ Battaglia et al. (2008)
- ↑ 5.0 5.1 ISOGG, Copyright 2016 by. "ISOGG 2017 Y-DNA Haplogroup E". https://isogg.org/tree/2017/ISOGG_HapgrpE17.html.
- ↑ E-CTS693
- ↑ 7.0 7.1 Ingman, Tara; Eisenmann, Stefanie; Skourtanioti, Eirini; Akar, Murat; Ilgner, Jana; Gnecchi Ruscone, Guido Alberto; le Roux, Petrus; Shafiq, Rula et al. (2021). "Human mobility at Tell Atchana (Alalakh), Hatay, Turkey during the 2nd millennium BC: Integration of isotopic and genomic evidence". PLOS ONE 16 (6): e0241883. doi:10.1371/journal.pone.0241883. PMID 34191795. Bibcode: 2021PLoSO..1641883I.
- ↑ 8.0 8.1 Antonio, Margaret L. et al. (8 November 2019). "Ancient Rome: A genetic crossroads of Europe and the Mediterranean". Science (American Association for the Advancement of Science) 366 (6466): 708–714. doi:10.1126/science.aay6826. PMID 31699931. Bibcode: 2019Sci...366..708A.
- ↑ Olalde, Iñigo; Carrión, Pablo; Mikić, Ilija; Rohland, Nadin; Mallick, Shop; Lazaridis, Iosif; Korać, Miomir; Golubović, Snežana et al. (2021-08-31) (in en). Cosmopolitanism at the Roman Danubian Frontier, Slavic Migrations, and the Genomic Formation of Modern Balkan Peoples. doi:10.1101/2021.08.30.458211. http://biorxiv.org/lookup/doi/10.1101/2021.08.30.458211.
- ↑ Beniamino Trombetta (2015). "Phylogeographic refinement and large scale genotyping of human Y chromosome haplogroup E provide new insights into the dispersal of early pastoralists in the African continent". Genome Biology and Evolution 7 (7): 1940–1950. doi:10.1093/gbe/evv118. PMID 26108492.
- ↑ Hassan et al. (2008)
- ↑ Hirbo, Jibril Boru. "Complex Genetic History of East African Human Populations". University of Maryland, College Park. http://drum.lib.umd.edu/bitstream/1903/11443/1/Hirbo_umd_0117E_11892.pdf.
Sources
- Battaglia, Vincenza; Fornarino, Simona; Al-Zahery, Nadia; Olivieri, Anna; Pala, Maria; Myres, Natalie M; King, Roy J; Rootsi, Siiri et al. (2008), "Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe", European Journal of Human Genetics 17 (6): 820–830, doi:10.1038/ejhg.2008.249, PMID 19107149
- Cruciani, Fulvio; La Fratta, Roberta; Santolamazza, Piero; Sellitto, Daniele; Pascone, Roberto; Moral, Pedro; Watson, Elizabeth; Guida, Valentina et al. (May 2004). "Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out Of Africa". The American Journal of Human Genetics 74 (5): 1014–1022. doi:10.1086/386294. PMID 15042509.
- Cruciani; La Fratta; Torroni; Underhill; Scozzari (2006), "Molecular Dissection of the Y Chromosome Haplogroup E-M78 (E3b1a): A Posteriori Evaluation of a Microsatellite-Network-Based Approach Through Six New Biallelic Markers", Human Mutation 27 (8): 831–2, doi:10.1002/humu.9445, PMID 16835895
- Cruciani, F.; La Fratta, R.; Trombetta, B.; Santolamazza, P.; Sellitto, D.; Colomb, E. B.; Dugoujon, J.-M.; Crivellaro, F. et al. (2007), "Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12", Molecular Biology and Evolution 24 (6): 1300–1311, doi:10.1093/molbev/msm049, PMID 17351267, http://mbe.oxfordjournals.org/cgi/reprint/24/6/1300
- Hassan, Hisham Y.; Underhill, Peter A.; Cavalli-Sforza, Luca L.; Ibrahim, Muntaser E. (2008), "Y-Chromosome Variation Among Sudanese: Restricted Gene Flow, Concordance With Language, Geography, and History", American Journal of Physical Anthropology 137 (3): 316–23, doi:10.1002/ajpa.20876, PMID 18618658, http://dirkschweitzer.net/E3b-papers/Hassan-Sudan-2008-AJPA.pdf
- Sanchez, Juan J; Hallenberg, Charlotte; Børsting, Claus; Hernandez, Alexis; Morling, Niels (9 March 2005). "High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males". European Journal of Human Genetics 13 (7): 856–866. doi:10.1038/sj.ejhg.5201390. PMID 15756297.
- Tillmar, Andreas O.; Montelius, Kerstin (December 2009). "Population data of 12 Y-STR loci from a Somali population". Forensic Science International: Genetics Supplement Series 2 (1): 413–415. doi:10.1016/j.fsigss.2009.08.078.
- Underhill, P. A.; Passarino, G.; Lin, A. A.; Shen, P.; Mirazon Lahr, M.; Foley, R. A.; Oefner, P. J.; Cavalli-Sforza, L. L. (January 2001). "The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations". Annals of Human Genetics 65 (1): 43–62. doi:10.1046/j.1469-1809.2001.6510043.x. PMID 11415522.
 |

