Biology:Haplogroup E-M215 (Y-DNA)
| Haplogroup E-M215 | |
|---|---|
| Possible time of origin | 47,500 - 22,400 years BP[1][2][3] |
| Possible place of origin | East Africa[citation needed] (Horn of Africa)[1][2] |
| Ancestor | E-P2 |
| Descendants | E-M35, E-M281 |
| Defining mutations | M215 |
E-M215, also known as E1b1b and formerly E3b, is a major human Y-chromosome DNA haplogroup. It is a division of the macro-haplogroup E-M96, which is defined by the single-nucleotide polymorphism (SNP) mutation M215.[4][5][6] In other words, it is one of the major patrilineages of humanity, linking from father-to-son back to a common male-line ancestor ("Y-chromosomal Adam"). It is a subject of discussion and study in genetics as well as genetic genealogy, archaeology, and historical linguistics.
The E-M215 haplogroup has two ancient branches that contain all the known modern E-M215, E-M35 and E-M281 subclades. Of the latter two E-M215 subhaplogroups, the only branch that has been confirmed in a native population outside of Ethiopia is E-M35. E-M35 in turn has two known branches, E-V68 and E-Z827, which contain by far the majority of all modern E-M215 subclade bearers. The E-V257 and E-V68 subclades each respectively their highest frequencies in North Africa and the Horn of Africa, and also at lower percentages in parts of the Middle East and Europe, and in isolated populations of Southern Africa.
Origins
The origins of E-M215 were dated by Cruciani in 2007 to about 22,400 years ago in the Horn of Africa.[3][Note 1] E-M35 was dated by Batini in 2015 to between 15,400 and 20,500 years ago.[7] In June 2015, Trombetta et al. reported a previously unappreciated large difference in the age between haplogroup E-M215 (38.6 kya; 95% CI 31.4–45.9 kya) and its sub-haplogroup E-M35 (25.0 kya; 95% CI 20.0–30.0 kya) and estimated its origin to be in Northeast Africa, where the node separating the E-V38 and E-M215 branches occurs about 47,500 years ago (95% CI: 41.3–56.8 ka).[1]
All major sub-branches of E-M35 are thought to have originated in the same general area as the parent clade: in North Africa, the Horn of Africa, or nearby areas of the Near East. Some branches of E-M35 are assumed to have left Africa thousands of years ago, whereas others may have arrived from the Near East. For example, Underhill (2002) associates the spread of the haplogroup with the Neolithic Revolution, believing that the structure and regional pattern of E-M35 subclades potentially give "reagents with which to infer specific episodes of population histories associated with the Neolithic agricultural expansion". Battaglia et al. (2007) also estimate that E-M78 (called E1b1b1a1 in that paper) has been in Europe longer than 10,000 years. Accordingly, human remains excavated in a Spanish funeral cave dating from approximately 7,000 years ago were shown to be in this haplogroup.[11] Two more E-M78 have been found in the Neolithic Sopot and Lengyel cultures too.[12]
Concerning E-M35 in Europe within this scheme, Underhill & Kivisild (2007) have remarked that E-M215 seems to represent a late-Pleistocene migration from North Africa to Europe over the Sinai Peninsula in Egypt.[Note 2] While this proposal remains uncontested, it has more recently been proposed by Trombetta et al. (2011) that there is also evidence for additional migration of E-M215 carrying men directly from North Africa to southwestern Europe, via a maritime route (see below.)
According to Lazaridis et al. (2016), Natufian skeletal remains from the ancient Levant predominantly carried the Y-DNA haplogroup E1b1b. Of the five Natufian specimens analysed for paternal lineages, three belonged to the E1b1b1b2(xE1b1b1b2a,E1b1b1b2b), E1b1(xE1b1a1,E1b1b1b1) and E1b1b1b2(xE1b1b1b2a,E1b1b1b2b) subclades (60%). Haplogroup E1b1b was also found at moderate frequencies among fossils from the ensuing Pre-Pottery Neolithic B culture, with the E1b1b1 and E1b1b1b2(xE1b1b1b2a,E1b1b1b2b) subclades observed in two of seven PPNB specimens (~29%). The scientists suggest that the Levantine early farmers may have spread southward into East Africa, bringing along Western Eurasian and Basal Eurasian ancestral components separate from that which would arrive later in North Africa.
Additionally, haplogroup E1b1b1 has been found in an ancient Egyptian mummy excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which dates from a period between the late New Kingdom and the Roman era.[13] Fossils at the Iberomaurusian site of Ifri n'Amr or Moussa in Morocco, which have been dated to around 5,000 BCE, also carried haplotypes related to the E1b1b1b1a (E-M81) subclade. These ancient individuals bore an autochthonous Maghrebi genomic component that peaks among modern North Africans, indicating that they were ancestral to populations in the area.[14] The E1b1b haplogroup has likewise been observed in ancient Guanche fossils excavated in Gran Canaria and Tenerife on the Canary Islands, which have been radiocarbon-dated to between the 7th and 11th centuries CE. The clade-bearing individuals that were analysed for paternal DNA were inhumed at the Tenerife site, with all of these specimens found to belong to the E1b1b1b1a1 or E-M183 subclade (3/3; 100%).[15]
Loosdrecht et al. (2018) analysed genome-wide data from seven ancient Iberomaurusian individuals from the Grotte des Pigeons near Taforalt in eastern Morocco. The fossils were directly dated to between 15,100 and 13,900 calibrated years before present. The scientists found that five male specimens with sufficient nuclear DNA preservation belonged to the E1b1b1a1 (M78) subclade, with one skeleton bearing the E1b1b1a1b1 parent lineage to E-V13, another male specimen belonged to E1b1b (M215*).[16]
Distribution
In Africa, E-M215 is distributed in highest frequencies in the Horn of Africa and North Africa, specifically in the countries Somalia and Morocco, whence it has in recent millennia expanded as far south as South Africa , and northwards into Western Asia and Europe (especially the Mediterranean and the Balkans).[8][17][18][19]
Almost all E-M215 men are also in E-M35. In 2004, M215 was found to be older than M35 when individuals were found who have the M215 mutation, but do not have M35 mutation.[8] In 2013, one individual in Khorasan, North-East Iran, was found by Di Cristofaro et al. (2013) to be positive for M215 but negative for M35.
E-M215 and E-M35 are quite common among Afroasiatic speakers. The linguistic group and carriers of E-M35 lineage have a high probability to have arisen and dispersed together from the Afroasiatic Urheimat.[20] Amongst populations with an Afro-Asiatic speaking history, a significant proportion of Jewish male lineages are E-M35.[21] Haplogroup E-M35, which accounts for approximately 18%[17] to 20%[22][23] of Ashkenazi and 8.6%[24] to 30%[17] of Sephardi Y-chromosomes, appears to be one of the major founding lineages of the Jewish population.[25][Note 3]
The following table only includes sample populations with more than 1% E-M215 men with all known subclades as of June 2015. It contains the E-V1515 clade defined by Trombetta et al. 2015, and all the E1b1b subclades distributed below the Sahara (E-V42, E-M293, E-V92, E-V6), which were identified as E-M35 basal clades in a former phylogeny.[1]
| Population | N | Region | Language | Total E-M215 | E-V2009 | E-M78* | E-V1477 | E-V1083* | E-V13 | E-V22 | E-V12* | E-V32 | E-V259 | E-V65 | E-V257* | E-M81 | E-M123* | E-M34 | E-V1515* | E-V1486* | E-V2881* | E-V1792 | E-V92 | E-M293* | E-V3065 | E-V42 | E-V1785* | E-V6 | E-V16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Northern Africa | |||||||||||||||||||||||||||||
| Moroccan Arabs | 55 | Morocco | AA/Semitic | 15.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.3 | 0.0 | 0.0 | 0.0 | 32.7 | 0.0 | 30.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Asni Berbers | 54 | Morocco | AA/Berber | 85.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.7 | 0.0 | 0.0 | 0.0 | 0.0 | 1.9 | 79.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Bouhria Berbers | 67 | Morocco | AA/Berber | 79.1 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 77.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Middle Atlas Berbers | 69 | Morocco | AA/Berber | 81.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.1 | 0.0 | 71.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Marrakech Berbers | 27 | Morocco | AA/Berber | 92.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.7 | 3.7 | 0.0 | 0.0 | 0.0 | 3.7 | 77.8 | 0.0 | 3.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Souss Berbers | 34 | Morocco | AA/Berber | 79.4 | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 76.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Ouarzazate Berbers | 31 | Morocco | AA/Berber | 54.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 54.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Mozabite Berbers | 67 | Algeria | AA/Berber | 89.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 0.0 | 86.6 | 0.0 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Tunisian Jews | 10 | Tunisia | AA/Semitic | 20.0 | 0.0 | 0.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Libyan Arabs | 10 | Libya | AA/Semitic | 20.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 | 30.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Libyan Jews | 23 | Libya | AA/Semitic | 26.1 | 0.0 | 0.0 | 0.0 | 0.0 | 4.3 | 0.0 | 0.0 | 0.0 | 0.0 | 4.3 | 0.0 | 0.0 | 0.0 | 17.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Northern Egyptians | 49 | Egypt | AA/Semitic | 20.9 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 16.3 | 4.1 | 2.0 | 0.0 | 0.0 | 0.0 | 4.1 | 4.1 | 10.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Egyptian Berbers from Siwa | 93 | Egypt | AA/Semitic | 18.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.2 | 0.0 | 0.0 | 4.3 | 2.2 | 1.1 | 0.0 | 2.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.5 | 0.0 |
| Egyptians from Baharia | 41 | Egypt | AA/Semitic | 56.1 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 | 22.0 | 14.6 | 0.0 | 0.0 | 2.4 | 7.3 | 4.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 |
| Egyptians from Gurna Oasis | 34 | Egypt | AA/Semitic | 17.6 | 0.0 | 5.9 | 0.0 | 0.0 | 0.0 | 0.0 | 8.8 | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Southern Egyptians | 47 | Egypt | AA/Semitic | 78.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 74.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.1 | 2.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Western/Central Africa | |||||||||||||||||||||||||||||
| Mandenka | 16 | Senegal | NC/Mande | 6.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Tuareg | 22 | Niger | AA/Berber | 13.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.5 | 0.0 | 0.0 | 0.0 | 0.0 | 9.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Daba | 29 | Cameroon (North) | AA/Chadic | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Guidar | 9 | Cameroon (North) | AA/Chadic | 11.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 11.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Mandara | 82 | Cameroon (North) | AA/Chadic | 2.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 | 0.0 | 0.0 | 0.0 | 1.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Shuwa Arabs | 5 | Cameroon (North) | AA/Semitic | 20.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Fulbe from Cameroon | 76 | Cameroon (North) | NC/Atlantic | 1.3 | 1.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Moundang | 21 | Cameroon (North) | NC/Adamawa | 4.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Eastern Africa | |||||||||||||||||||||||||||||
| Tigre | 5 | Eritrea | AA/Semitic | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 60.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 |
| Nara | 15 | Eritrea | NS/Sudanic | 60.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.7 | 0.0 | 13.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 13.3 | 6.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 13.3 | 0.0 | 6.7 | 0.0 |
| Cunama | 20 | Eritrea | NS/Cunama | 65.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 20.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 15.0 | 10.0 | 5.0 | 0.0 |
| Saho | 94 | Eritrea | AA/Cushitic | 98.9 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | 88.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.5 | 0.0 |
| Tigrai | 32 | Eritrea/Ethiopia | AA/Semitic | 71.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.1 | 3.1 | 21.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.1 | 3.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 31.3 | 6.3 | 0.0 |
| Afar | 25 | Djibouti | AA/Cushitic | 60.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 56.0 | 0.0 |
| Somali | 40 | Djibouti | AA/Cushitic | 25.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 25.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Ethiopian Jews | 22 | Ethiopia | AA/Cushitic | 31.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 13.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.1 | 0.0 | 0.0 | 0.0 |
| Amhara | 82 | Ethiopia | AA/Semitic | 45.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 | 11.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 13.4 | 0.0 | 0.0 | 2.4 | 0.0 | 1.2 | 0.0 | 0.0 | 1.2 | 0.0 | 8.5 | 4.9 |
| Oromo | 62 | Ethiopia | AA/Cushitic | 53.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 22.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.8 | 0.0 | 0.0 | 17.7 | 0.0 | 0.0 | 1.6 | 0.0 | 3.2 | 0.0 | 1.6 | 1.6 |
| Wolayta | 12 | Ethiopia | AA/Omotic | 58.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.3 | 0.0 | 8.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.3 | 0.0 | 0.0 | 8.3 | 0.0 | 0.0 | 8.3 | 0.0 | 0.0 | 0.0 | 16.7 | 0.0 |
| Somali | 12 | Ethiopia | AA/Cushitic | 50.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.3 | 0.0 | 25.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Gurage | 7 | Ethiopia | AA/Semitic | 42.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 28.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 14.3 | 0.0 |
| Somali | 5 | Somalia | AA/Cushitic | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 80.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Turkana | 6 | Kenya | NS/Sudanic | 50.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 33.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Borana | 9 | Kenya | AA/Cushitic | 77.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 66.7 | 0.0 | 0.0 | 11.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Somali | 6 | Kenya | AA/Cushitic | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 66.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.7 | 0.0 |
| Nilotic Western Kenya | 11 | Kenya | NS/Sudanic | 45.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.1 | 9.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 18.2 | 0.0 | 0.0 | 0.0 | 9.1 | 0.0 |
| Luhya | 51 | Kenya | NC/Bantu | 9.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Other Bantu | 17 | Kenya | NC/Bantu | 11.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 11.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Kikuyu | 9 | Kenya | NC/Bantu | 11.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 11.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Maasai | 45 | Kenya | NS/Sudanic | 37.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.7 | 0.0 | 6.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 24.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Tutsi | 9 | Burundi | NC/Bantu | 22.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 22.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Southern Africa | |||||||||||||||||||||||||||||
| !Kung | 64 | Angola | KS | 10.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.4 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 |
| Khwe | 26 | Namibia | KS | 30.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 30.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Bantu | 8 | South Africa | NC/Bantu | 12.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Europe | |||||||||||||||||||||||||||||
| Northern Portuguese | 50 | Portugal | IE | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Southern Portuguese | 49 | Portugal | IE | 16.3 | 0.0 | 0.0 | 0.0 | 0.0 | 4.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Pasiegos from Cantabria | 56 | Spain | IE | 42.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.8 | 41.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Asturians | 90 | Spain | IE | 12.2 | 0.0 | 0.0 | 0.0 | 0.0 | 5.6 | 4.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Southern Spaniards | 62 | Spain | IE | 6.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.2 | 0.0 | 0.0 | 0.0 | 0.0 | 1.6 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Spanish Basques | 55 | Spain | Basque | 3.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| French | 85 | France | IE | 8.2 | 0.0 | 0.0 | 0.0 | 0.0 | 3.5 | 0.0 | 1.2 | 0.0 | 0.0 | 0.0 | 0.0 | 3.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| French Basques | 16 | France | Basque | 6.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Corsicans | 140 | France | IE | 6.4 | 0.0 | 0.0 | 0.0 | 0.0 | 4.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.7 | 0.0 | 0.0 | 1.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Danish | 35 | Denmark | IE | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Germans | 77 | Germany | IE | 3.9 | 0.0 | 0.0 | 0.0 | 0.0 | 3.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Northern Italians | 80 | Italy | IE | 11.3 | 0.0 | 0.0 | 0.0 | 0.0 | 6.3 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | 0.0 | 1.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Central Italians | 356 | Italy | IE | 12.9 | 0.0 | 0.0 | 0.0 | 0.0 | 5.3 | 2.0 | 0.3 | 0.0 | 0.0 | 0.3 | 0.3 | 0.8 | 0.0 | 3.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Southern Italians | 141 | Italy | IE | 15.6 | 0.7 | 0.0 | 0.0 | 0.0 | 8.5 | 1.4 | 0.7 | 0.0 | 0.0 | 0.0 | 0.0 | 1.4 | 0.0 | 2.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Sicilians | 153 | Italy | IE | 20.3 | 0.0 | 0.0 | 0.0 | 0.0 | 7.2 | 4.6 | 0.7 | 0.0 | 0.0 | 0.7 | 0.0 | 0.7 | 0.0 | 6.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Sardinians | 374 | Italy | IE | 8.3 | 0.8 | 0.0 | 0.0 | 0.3 | 1.1 | 0.8 | 0.3 | 0.0 | 0.0 | 1.1 | 0.3 | 0.3 | 0.0 | 3.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Polish | 40 | Poland | IE | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Slovenians | 104 | Slovenia | IE | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Estonians | 74 | Estonia | U | 5.4 | 0.0 | 0.0 | 0.0 | 0.0 | 4.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Hungarians | 106 | Hungary | U | 10.4 | 0.0 | 0.0 | 0.0 | 0.0 | 9.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Romanians | 30 | Romania | IE | 26.7 | 0.0 | 0.0 | 0.0 | 0.0 | 26.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Macedonians | 99 | Macedonia | IE | 18.2 | 0.0 | 0.0 | 0.0 | 0.0 | 18.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Continental Greeks | 32 | Greece | IE | 28.1 | 0.0 | 0.0 | 0.0 | 0.0 | 25.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Bulgarians | 112 | Bulgaria | IE | 22.3 | 0.0 | 0.0 | 0.0 | 0.0 | 21.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Sephardic Bulgarians | 20 | Bulgaria | IE | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Albanians | 21 | Albania | IE | 33.3 | 0.0 | 0.0 | 0.0 | 0.0 | 33.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Near East | |||||||||||||||||||||||||||||
| Sephardic Turkish | 19 | Turkey | A | 10.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.3 | 0.0 | 5.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Istanbul Turkish | 35 | Turkey | A | 17.1 | 0.0 | 0.0 | 0.0 | 0.0 | 2.9 | 5.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.7 | 0.0 | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Southwestern Turkish | 40 | Turkey | A | 7.5 | 0.0 | 0.0 | 0.0 | 0.0 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.5 | 0.0 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Northeastern Turkish | 41 | Turkey | A | 2.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Central Anatolian | 61 | Turkey | A | 9.8 | 0.0 | 0.0 | 0.0 | 0.0 | 4.9 | 0.0 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Southeastern Turkish | 24 | Turkey | A | 8.3 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Erzurum Turkish | 25 | Turkey | A | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Turkish Cypriots | 46 | Turkey | A | 23.9 | 0.0 | 0.0 | 0.0 | 0.0 | 10.9 | 2.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.7 | 0.0 | 2.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Bedouins | 28 | Israel | AA/Semitic | 14.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.6 | 0.0 | 7.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Druze Arabs | 28 | Israel | AA/Semitic | 14.3 | 0.0 | 0.0 | 0.0 | 0.0 | 10.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Palestinians | 29 | Israel | AA/Semitic | 13.8 | 0.0 | 0.0 | 0.0 | 0.0 | 3.4 | 6.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Arabs | 41 | United Arab Emirates | AA/Semitic | 7.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Omanite | 13 | Oman | AA/Semitic | 15.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Yemenites | 94 | Yemen | AA/Semitic | 14.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.1 | 3.2 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | 7.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 |
E-M215 association with endurance
Moran et al. (2004) observed that among Y-DNA (paternal) clades borne by elite endurance athletes in Ethiopia, the haplogroup E3b1 was negatively correlated with elite athletic endurance performance,[26] whereas the haplogroups E*, E3*, K*(xP),[26] and J*(xJ2) were significantly more frequent among the elite endurance athletes.[26]
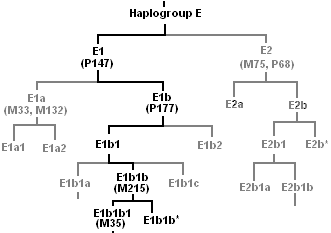
Subclades of E-M215
Cladogram with the main subclades:
| E1b1b (M215) |
| ||||||||||||||||||||||||||||||||||||
Family tree
The following phylogenetic tree is based on the YCC 2008 tree and subsequent published research as summarized by ISOGG. It includes all known subclades as of June 2015 (Trombetta et al. 2015)[27][5][6]
- E-M215 (E1b1b)
- E-M215*. Rare or non-existent.
- E-M35 (E1b1b1)
- E-V68 (E1b1b1a)
- E-V2009. Found in individuals in Sardinia and Morocco.
- E-M78 (E1b1b1a1). North Africa, Horn of Africa, West Asia, Sicily. (Formerly "E1b1b1a".)
- E-M78*
- E-V1477. Found in Tunisian Jews.
- E-V1083.
- E-V1083*. Found only in Eritrea (1.1%) and Sardinia (0.3%).
- E-V13
- E-V22
- E-V1129
- E-V12
- E-V12*
- E-V32
- E-V264
- E-V259. Found in North Cameroon.
- E-V65
- E-CTS194
- E-V12
- E-Z827 (E1b1b1b)[28]
- E-V257/L19 (L19, V257) – E1b1b1b1[28]
- E-PF2431
- E-M81 (M81)
- E-PF2546
- E-PF2546*
- E-CTS12227
- E-MZ11
- E-MZ12
- E-MZ11
- E-A929
- E-Z5009
- E-Z5009*
- E-Z5010
- E-Z5013
- E-Z5013*
- E-A1152
- E-A2227
- E-A428
- E-MZ16
- E-PF6794
- E-PF6794*
- E-PF6789
- E-MZ21
- E-MZ23
- E-MZ80
- E-A930
- E-Z2198/E-MZ46
- E-A601
- E-L351
- E-Z5009
- E-PF2546
- E-Z830 (Z830) – E1b1b1b2[28]
- E-M123 (M123)
- E-M34 (M34)
- E-M84 (M84)
- E-M136 (M136)
- E-M290 (M290)
- E-V23 (V23)
- E-L791 (L791,L792)
- E-M84 (M84)
- E-M34 (M34)
- E-V1515. E-V1515 and its subclades are mainly restricted to eastern Africa.
- E-V1515*
- E-V1486
- E-V1486*
- E-V2881
- E-V2881*
- E-V1792
- E-V92
- E-M293 (M293)
- E-M293*
- E-P72 (P72)
- E-V3065*
- E-V1700
- E-V42 (V42)
- E-V1785
- E-V1785*
- E-V6 (V6)
- E-M123 (M123)
- E-V257/L19 (L19, V257) – E1b1b1b1[28]
- E-V16/E-M281 (E1b1b2). Rare. Found in individuals in Ethiopia, Yemen and Saudi Arabia.
- E-V68 (E1b1b1a)
Exceptional cases of men who are M215 positive but M35 negative ("E-M215*") have been discovered so far in two Amharas of Ethiopia and one Yemeni.[8][29] At least some of these men, perhaps all, are known since early 2011 to be in a rare sibling clade to E-M35, known as E-V16 or E-M281.[30] The discovery of M281 was announced by Semino et al. (2002), who found it in two Ethiopian Oromo. Trombetta et al. (2011) found 5 more Ethiopian individuals and an equivalent SNP to M281, V16. It was in the 2011 paper that the family tree position (M215+/M35-) was discovered as described above. The E-M215 derivative, E-M35 is defined by the M35 SNP. 1 Turkmen individual from Jawzjan with a subclade defining mutation is referred to as E-M35*.[31] As of June 2015, there is an increasingly complex tree structure which divides most men in E-M35 into two branches: E-V68 and E-Z827.
The most frequently described subclades are E-M78, a part of E-V68, and E-M81, which is a branch of E-Z827. These two subclades represent the largest proportion of the modern E-M215 population. E-M78 is found over most of the range where E-M215 is found excluding Southern Africa. E-M81 is found mainly in North Africa. E-M123 is less common but widely scattered, with significant populations in specific parts of the Horn of Africa, the Levant, Arabia, Iberia, and Anatolia. A new clade (E-V1515) was defined by Trombetta et al. 2015, which originated about 12 kya (95% CI 8.6–16.4) in eastern Africa where it is currently mainly distributed. This clade includes the E-V42, E-M293, E-V92 and E-V6 subclades, which were identified as E-M35 basal clades in a previous phylogeny.[1]
Within E-M35, there are striking parallels between two haplogroups, E-V68 and E-V257. Both contain a lineage which has been frequently observed in Africa (E-M78 and E-M81, respectively) and a group of undifferentiated chromosomes that are mostly found in southern Europe. An expansion of E-M35 carriers, possibly from the Middle East as proposed by other authors, and split into two branches separated by the geographic barrier of the Mediterranean Sea, would explain this geographic pattern. However, the absence of E-V68* and E-V257* in the Middle East makes a maritime spread between northern Africa and southern Europe a more plausible hypothesis.
TMRCA of the major nodes in E-M215
| TMRCA (kya) | Trombetta 2015 | YFull |
| E-M215 | 39 | 35,4 |
| *E-M35 | 25 | 23,9 |
| **E-V68 | 20 | 20 |
| ***E-M78 | 15 | 13 |
| **E-Z827 | ? | 23,6 |
| ***E-V257/L19 | ? | 13,9 |
| ****E-M81 | ? | 2,7 |
| ***E-Z830 | 20 | 19 |
| ****E-M34 | ? | 15 |
| ****E-V1515 | 19 | ? |
E-V68 (E1b1b1a)
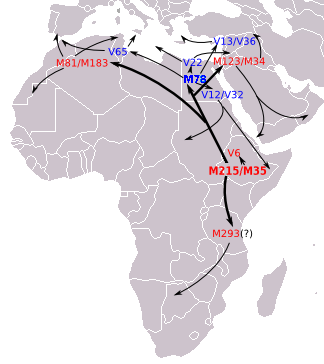
E-V68, is dominated by its longer-known subclade E-M78. Three "E-V68*" individuals who are in E-V68 but not E-M78 have been reported in Sardinia, by Trombetta et al. (2011), when announcing the discovery of V68. The authors noted that because E-V68* was not found in the Middle Eastern samples, this appears to be evidence of maritime migration from Africa to southwestern Europe. E-M78 is a commonly occurring subclade, widely distributed in North Africa, the Horn of Africa, West Asia, (the Middle East and Near East) "up to Southern Asia",[3] and all of Europe.[32] The European distribution has a frequency peak centered in parts of the Balkans (up to almost 50% in some areas)[17][33] and Sicily, and declining frequencies evident toward western, central, and northeastern Europe.
Based on genetic STR variance data, Cruciani et al. (2007) suggests that E-M78 originated in the region of Egypt and Libya.[Note 4] about 18,600 years ago (17,300 – 20,000 years ago).[Note 5] Battaglia et al. (2008) describe Egypt as "a hub for the distribution of the various geographically localized M78-related subclades" and, based on archaeological data, they propose that the point of origin of E-M78 (as opposed to later dispersal from Egypt) may have been in a refugium which "existed on the border of present-day Sudan and Egypt, near Lake Nubia, until the onset of a humid phase around 8500 BC. The northward-moving rainfall belts during this period could have also spurred a rapid migration of Mesolithic foragers northwards in Africa, the Levant and ultimately onward to Asia Minor and Europe, where they each eventually differentiated into their regionally distinctive branches". Towards the south, Hassan et al. (2008) also explain evidence that some subclades of E-M78, specifically E-V12 and E-V22, "might have been brought to Sudan from North Africa after the progressive desertification of the Sahara around 6,000–8,000 years ago". And similarly, Cruciani et al. (2007) propose that E-M78 in Ethiopia, Somalia and surrounding areas, back-migrated to this region from the direction of Egypt after acquiring the E-M78 mutation.
Recently, E-M78 was dated by Trombetta et al. 2015. between 20,300 and 14,800 years ago.[1]
Subclades of E-M78
Listed here are the main subclades of M78 as of June 2015. Within the E-M78 subclade, Trombetta et al. 2015 allocated most of the former E-M78* chromosomes to three new distinct branches: E-V1083*, E-V1477 and E-V259. The first is a paragroup sister to clades E-V22 and E-V13. The mutation V1477 defines a new basal branch that has been observed only in one northern African sample. Finally, a sister clade of E-V12 defined by V264 includes E-V65 and V259, a new lineage distributed in central Africa.[1][32]
- E-M78 (E1b1b1a1) North Africa, Horn of Africa, West Asia, Europe (formerly "E1b1b1a").
- E-M78* Found in Morocco, southern Portugal, southern Spain and Iran (Tehran and Semnan provinces).
- E-V1477 Found in Tunisian Jews.
- E-V1083
- E-V1083* Found only in Eritrea (1.1%) and Sardinia (0.3%).
- E-V13 This is the most common subclade of E-M215 found in Europe. It is especially common in the Balkans.
- E-V22. Concentrated in Northeast Africa and the Near East. Peaks among the Saho.
- E-V1129
- E-V12. Found in Egypt, Sudan, and Chad other places. Has an important subclade
- E-V12* Most common lineage among Southern Egyptians (74.5%).
- E-V32. Very common among Somalis, Tigre and Oromos.
- E-V264
- E-V259 Found in North Cameroon.
- E-V65 Associated with North Africa, but also found in Sicily and also found in continental Italy.
- E-V12. Found in Egypt, Sudan, and Chad other places. Has an important subclade
- E-M521 Not mentioned by Trombetta et al.2015. Found in two individuals in Greece by Battaglia et al. (2008) and in one individual from the Eastern Alpine region of Italy by Coia et al. (2013)
E-Z827 (E1b1b1b)
In human genetics, E-Z827, is the name of a major human Y-chromosome DNA haplogroup abundantly found in North Africa, particularly the Maghreb, and to a lesser extent in Horn of Africa, the Near East and Europe.
E-V257/E-L19 (E1b1b1b1)
E-V257* individuals in their samples who were E-V257, but not E-M81. A Borana from Kenya, a Marrakesh Berber, a Corsican, a Sardinian, a southern Spaniard and a Cantabrian. As mentioned above, Trombetta et al. (2011) propose that the absence of E-V257* in the Middle East (Yfull found a young one in Iranian Azerbaijan[34] and a different young one in Armenia[35]) makes a maritime movement from northern Africa to southern Europe the most plausible hypothesis so far to explain its distribution. Yfull lists 24 individuals, all of whom belong to a single branch that is 30% younger than their common ancestor with M81.[36]
E-M81
E-M81 is the most common subclade of haplogroup E-L19/V257. It is concentrated in the Maghreb, and is dominated by its E-M183 subclade. E-M183 is believed to have originated in the Northwest Africa and has an estimated age of 4700 ybp.[37]
This haplogroup reaches a mean frequency of 61% in the Maghreb and 51% in North Africa, decreasing in frequency from approximately 80% to 100% in Berber populations,[38][39] including Saharawis, to approximately 29% to the east of this range in Egypt.[40][41][42][43] Because of its young age and prevalence among these groups and also others such as Mozabite, Middle Atlas, Kabyle and other Berber groups, it is sometimes referred to as a genetic "Berber marker". Pereira et al. (2010) report high levels amongst Tuareg in two Saharan populations – 77.8% near Gorom-Gorom, in Burkina Faso, and 81.8% from Gosi in Mali. There was a much lower frequency of 11.1% in the vicinity of Tanut in Niger. E-M81 is also quite common among Maghrebi Arabic-speaking groups. It is generally found at frequencies around 45% in coastal cities of Algeria and Tunisia (Jijel, Oran, Tizi Ouzou, Algiers, Tunis, Sousse).[40][44]
In this key area from Egypt to the Atlantic Ocean, Solé-Morata et al. (2017) report a M183-SM001 pattern of decreasing microsatellite haplotype variation (implying greater lineage age in the former areas) from the Reguibat tribe in Oran and they found M183* (not SM001) in Iberia, Libya and Morocco. Arredi et al. (2004) however showed microsatellite variation decrease from East to West, accompanied by a substantial increasing frequency. At the eastern extreme of this core range, Kujanova et al. (2009) found M81 in 28.6% (10 out of 35 men) in el-Hayez in the Libyan Desert in Egypt.
Arredi et al. (2004) believe the pattern of distribution and variance to be consistent with the hypothesis of a "demic diffusion" from the East. There is no autochthonous presence of E-M81 in the Near East (there is one in Lebanon[45]), indicating that M81 most likely emerged from its parent clade M35 either in North Africa, or possibly as far south as the Horn of Africa.[46]
In Europe, E-M81 is widespread but rare, in the Iberian Peninsula shows an average frequency of 4% (45/1140) in the Iberian Peninsula with frequencies reaching 3.5% in Galicia, 4% in Western Andalusia and Northwest Castile. However this study includes 153 individuals from Majorca, Minorca and Ibiza islands as well as 24 individuals from Gascony which are not in the Iberian Peninsula. Without these 177 individuals, average for Iberian Peninsula is 4.1% (40/963),[47] it is found at comparable levels to E-M78, with an average frequency of around 5%. Its frequencies are higher in the western half of the peninsula with frequencies reaching 8% in Extremadura and southern Portugal, 4% to 5% in Galicia, 5% in western Andalusia and 4% in northwest Castile and 9% to 17% in Cantabria.[24][48][49][50][51] The highest frequencies of this clade found so far in Europe were observed in the Valles Pasiegos from Cantabria, ranging from 5.5% (8/45)[51] to 41% (23/56).[8] An average frequency of 8.28% (54/652) has also been reported in the Spanish Canary Islands with frequencies over 10% in the three largest islands of Tenerife (10.68%), Gran Canaria (11.54%) and Fuerteventura (13.33%).[52]
E-M81 is also found in France ,[8] 2.70% (15/555) overall with frequencies surpassing 5% in Auvergne (5/89) and Île-de-France (5/91),[53][54] 0,7% to 5,8% in Sardinia,[55][56] approximately 2.12% overall in Sicily (but up to 7.14% in Piazza Armerina),[57] and in very much lower frequency near Lucera (1.7%), in continental Italy,[50] possibly due to ancient migrations during the Islamic, Roman, and Carthaginian empires. In a 2014 study by Stefania Sarno et al. with 326 samples from Cosenza, Reggio Calabria, Lecce and five Sicilian provinces, E-M81 shows an average frequency of 1.53%, but the typical Maghrebin core haplotype 13-14-30-24-9-11-13 has been found in only two out of the five E-M81 individuals. These results, along with the negligible contribution from North-African populations revealed by the admixture-like plot analysis, suggest only a marginal impact of trans-Mediterranean gene flows on the current SSI genetic pool.[57][58]
As a result of its old world distribution, this subclade is found throughout Brasil[59] 5.4% in Brazil (Rio de Janeiro),[Note 6] and among Hispanic men from California and Hawaii 2.4%.[60]
In smaller numbers, E-M81 men can be found in areas in contact with North Africa, both around the Sahara, in places like Sudan, and around the Mediterranean in places like Lebanon, Turkey, and amongst Sephardi Jews.
There are two recognized subclades of E-M81, although one is much more important than the other.[which?]
The E-M81 subclade has been found in ancient Guanche (Bimbapes) fossils excavated in Punta Azul, El Hierro, Canary Islands, which are dated to the 10th century (~44%).[61]
E-M107
Underhill (2000) found one example of E-M107 in Mali.
E-M183
E-M183 is extremely dominant (more than 99%[62]) within E-M81. Karafet et al. (2008) first described it as a subclade of E-M81. The known subclades of E-M183 include:
- E-M165 Underhill et al. (2000) found one example in Middle East.
- E-L351 Found in two related participants in The E-M35 Phylogeny Project.
E-Z830 (E1b1b1b2)
This is a recently discovered subclade which has not yet been included in most haplogroup trees, E-Z830 includes the confirmed subclades of E-M123, E-V1515 (E-M293, E-V42, E-V6, E-V92), and E-Z830*, and is a sibling clade to E-L19. Currently, [63] the E-M35 phylogeny project] recognizes four distinct clusters of Z830* carriers, two of which are exclusively Jewish in origin. The remaining two are significantly smaller, and include scattered individuals in Germany, Spain, Latin America, Egypt, and Ethiopia.[64][65][66][67]
E-M123
E-M123 is mostly known for its major subclade E-M34, which dominates this clade.[Note 7]
E-V1515
A new clade (E-V1515) was defined by Trombetta et al. 2015, which originated about 12 kya (95% CI 8.6–16.4) in eastern Africa where it is currently mainly distributed. This clade includes all the sub-Saharan haplogroups (E-V42, E-M293, E-V92, E-V6) reported as E-M35 basal clades in a previous phylogeny.[1]
E-M293
E-M293 is a subclade of E-V1515. It was first identified by ISOGG as the second clade within E-Z830. It was discovered before E-Z830, being announced in Henn (2008), which associated it with the spread of pastoralism from East Africa into Southern Africa. So far high levels have been found in specific ethnic groups in Tanzania and Southern Africa. Highest were the Datooga (43%), Khwe (Kxoe) (31%), Burunge (28%), and Sandawe peoples (24%). Henn (2008) in their study also found two Bantu-speaking Kenyan males with the M293 mutation.[9]
Other E-M215 subclades are rare in Southern Africa. The authors state "Without information about M293 in the Maasai, Hema, and other populations in Kenya, Sudan, and Ethiopia, we cannot pinpoint the precise geographic source of M293 with greater confidence. However, the available evidence points to present-day Tanzania as an early and important geographic locus of M293 evolution.". They also say that "M293 is only found in sub-Saharan Africa, indicating a separate phylogenetic history for M35.1 * (former) samples further north".
E-P72 appears in Karafet (2008). Trombetta et al. (2011) announced that this is a subclade of E-M293.
E-V42
Trombetta et al. (2011) announced the discovery of E-V42 in two Beta Israel persons. It was suggested that it may be restricted to the region around Ethiopia. However, further testing by commercial DNA testing companies confirmed many positive results for this subclade in Saudi Arabia, Kuwait and one person in Portugal who has a root from Arabia.[68]
E-V6
The E-V6 subclade of E-V1515 is defined by V6. Cruciani et al. (2004) identified a significant presence of these lineages in Ethiopia and also some in the neighboring Somalis. Among the Ethiopian and Somali samples, the highest were 14.7% among the Amhara and 16.7% among the Wolayta.
To the south, (Tishkoff Gonder) identified one V6+ man in a sample of 35 Datooga of Tanzania. And further to the north, (Dugoujon Coudray) identified another 6 men in a sample of 93 from the Siwa Oasis, which is a Berber population
E-V92
Trombetta et al. (2011) announced the discovery of E-V92 in two Amharas. Like E-V6 and E-V42 it possibly only exists in the area of Ethiopia.
Phylogenetics
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E-P29 | 21 | III | 3A | 13 | Eu3 | H2 | B | E* | E | E | E | E | E | E | E | E | E | E |
| E-M33 | 21 | III | 3A | 13 | Eu3 | H2 | B | E1* | E1 | E1a | E1a | E1 | E1 | E1a | E1a | E1a | E1a | E1a |
| E-M44 | 21 | III | 3A | 13 | Eu3 | H2 | B | E1a | E1a | E1a1 | E1a1 | E1a | E1a | E1a1 | E1a1 | E1a1 | E1a1 | E1a1 |
| E-M75 | 21 | III | 3A | 13 | Eu3 | H2 | B | E2a | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 |
| E-M54 | 21 | III | 3A | 13 | Eu3 | H2 | B | E2b | E2b | E2b | E2b1 | - | - | - | - | - | - | - |
| E-P2 | 25 | III | 4 | 14 | Eu3 | H2 | B | E3* | E3 | E1b | E1b1 | E3 | E3 | E1b1 | E1b1 | E1b1 | E1b1 | E1b1 |
| E-M2 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a* | E3a | E1b1 | E1b1a | E3a | E3a | E1b1a | E1b1a | E1b1a | E1b1a1 | E1b1a1 |
| E-M58 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a1 | E3a1 | E1b1a1 | E1b1a1 | E3a1 | E3a1 | E1b1a1 | E1b1a1 | E1b1a1 | E1b1a1a1a | E1b1a1a1a |
| E-M116.2 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a2 | E3a2 | E1b1a2 | E1b1a2 | E3a2 | E3a2 | E1b1a2 | E1b1a2 | E1ba12 | removed | removed |
| E-M149 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a3 | E3a3 | E1b1a3 | E1b1a3 | E3a3 | E3a3 | E1b1a3 | E1b1a3 | E1b1a3 | E1b1a1a1c | E1b1a1a1c |
| E-M154 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a4 | E3a4 | E1b1a4 | E1b1a4 | E3a4 | E3a4 | E1b1a4 | E1b1a4 | E1b1a4 | E1b1a1a1g1c | E1b1a1a1g1c |
| E-M155 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a5 | E3a5 | E1b1a5 | E1b1a5 | E3a5 | E3a5 | E1b1a5 | E1b1a5 | E1b1a5 | E1b1a1a1d | E1b1a1a1d |
| E-M10 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a6 | E3a6 | E1b1a6 | E1b1a6 | E3a6 | E3a6 | E1b1a6 | E1b1a6 | E1b1a6 | E1b1a1a1e | E1b1a1a1e |
| E-M35 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b* | E3b | E1b1b1 | E1b1b1 | E3b1 | E3b1 | E1b1b1 | E1b1b1 | E1b1b1 | removed | removed |
| E-M78 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b1* | E3b1 | E1b1b1a | E1b1b1a1 | E3b1a | E3b1a | E1b1b1a | E1b1b1a | E1b1b1a | E1b1b1a1 | E1b1b1a1 |
| E-M148 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b1a | E3b1a | E1b1b1a3a | E1b1b1a1c1 | E3b1a3a | E3b1a3a | E1b1b1a3a | E1b1b1a3a | E1b1b1a3a | E1b1b1a1c1 | E1b1b1a1c1 |
| E-M81 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2* | E3b2 | E1b1b1b | E1b1b1b1 | E3b1b | E3b1b | E1b1b1b | E1b1b1b | E1b1b1b | E1b1b1b1 | E1b1b1b1a |
| E-M107 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2a | E3b2a | E1b1b1b1 | E1b1b1b1a | E3b1b1 | E3b1b1 | E1b1b1b1 | E1b1b1b1 | E1b1b1b1 | E1b1b1b1a | E1b1b1b1a1 |
| E-M165 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2b | E3b2b | E1b1b1b2 | E1b1b1b1b1 | E3b1b2 | E3b1b2 | E1b1b1b2a | E1b1b1b2a | E1b1b1b2a | E1b1b1b2a | E1b1b1b1a2a |
| E-M123 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b3* | E3b3 | E1b1b1c | E1b1b1c | E3b1c | E3b1c | E1b1b1c | E1b1b1c | E1b1b1c | E1b1b1c | E1b1b1b2a |
| E-M34 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b3a* | E3b3a | E1b1b1c1 | E1b1b1c1 | E3b1c1 | E3b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1b2a1 |
| E-M136 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3ba1 | E3b3a1 | E1b1b1c1a | E1b1b1c1a1 | E3b1c1a | E3b1c1a | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1b2a1a1 |
Research publications
The following research teams per their publications were represented in the creation of the YCC Tree.
Discussion
E-M215 and E1b1b1 are the currently accepted names found in the proposals of the Y Chromosome Consortium (YCC), for the clades defined by mutation M215 and M35 respectively, which can also be referred to as E-M215 and E-M35.[5] The nomenclature E3b (E-M215) and E3b1 (E-M35) respectively were the YCC defined names used to designate the same haplogroups in older literature with E-M35 branching as a separate subclade of E-M215 in 2004.[8] Prior to 2002 these haplogroups were not designated in a consistent way, and nor was their relationship to other related clades within haplogroup E and haplogroup DE. But in non-standard or older terminologies, E-M215 is for example approximately the same as "haplotype V", still used in publications such as Gérard et al. (2006).[6]
See also
Genetics
Y-DNA E subclades
Y-DNA backbone tree
Notes
- ↑ Cruciani et al. (2004): "Several observations point to eastern Africa as the homeland for haplogroup E3b—that is, it had (1) the highest number of different E3b clades (table 1), (2) a high frequency of this haplogroup and a high microsatellite diversity, and, finally, (3) the exclusive presence of the undifferentiated E3b* paragroup." As mentioned above, "E3b" is the old name for E-M215. Semino et al. (2004): "This inference is further supported by the presence of additional Hg E lineal diversification and by the highest frequency of E-P2* and E-M35* in the same region. The distribution of E-P2* appears limited to eastern African peoples. The E-M35* lineage shows its highest frequency (19.2%) in the Ethiopian Oromo but with a wider distribution range than E-P2*." For E-M215 Cruciani et al. (2007) reduced their estimate to 22,400 from 25,600 in Cruciani et al. (2004), re-calibrating the same data.
- ↑ "Y chromosome data show a signal for a separate late-Pleistocene migration from Africa to Europe via Sinai as evidenced through the distribution of haplogroup E3b lineages, which is not manifested in mtDNA haplogroup distributions."Underhill & Kivisild (2007:547)
- ↑ "Paragroup E-M35 * and haplogroup J-12f2a* fit the criteria for major AJ founding lineages because they are widespread both in AJ populations and in Near Eastern populations, and occur at much lower frequencies in European non-Jewish populations." Behar et al. (2004)
- ↑ Cruciani et al. (2007) use the term Northeastern Africa to refer to Egypt and Libya, as shown in Table 1 of the study. Prior to Cruciani et al. (2007), Semino et al. (2004) East Africa as a possible place of origin of E-M78, based upon Ethiopian testing. This was because of the high frequency and diversity of E-M78 lineages in the region of Ethiopia. However, Cruciani et al. (2007) were able to study more data, including populations from North Africa who were not represented in the Semino et al. (2004) study, and found evidence that the E-M78 lineages which make up a significant proportion of some populations in that region, were relatively young branches (see E-V32 below). They therefore concluded that "Northeast Africa" was the likely place of origin of E-M78 based on "the peripheral geographic distribution of the most derived subhaplogroups with respect to northeastern Africa, as well as the results of quantitative analysis of UEP and microsatellite diversity". So according to Cruciani et al. (2007) E-M35, the parent clade of E-M78, originated in East Africa, subsequently spread to Northeast Africa, and then there was a "back migration" of E-M215 chromosomes that had acquired the E-M78 mutation. Cruciani et al. (2007) therefore note this as evidence for "a corridor for bidirectional migrations" between Northeast Africa (Egypt and Libya in their data) on the one hand and East Africa on the other. The authors believe there were "at least 2 episodes between 23.9–17.3 ky and 18.0–5.9 ky ago".
- ↑ Cruciani et al. (2007) use two calculation methods for estimating the age of E-M78 which give very different results. For the main 18,600 years ago, the ASD method is used, while for a second "ρ method", used as a check, gives 13.7kya with a standard deviation of 2.3kya, but the difference between the two methods is only large for the age estimation of E-M78, not its subclades. The authors state that the big difference is "attributable to the relevant departure from a star-like structure because of repeated founder effects"
- ↑ (6 out of 112), "The presence of chromosomes of North African origin (E3b1b-M81; Cruciani et al., 2004) can also be explained by a Portuguese but especially Italian-mediated influx, since this haplogroup reaches a frequency of 4.6% in Portugal and of 4.8% Italy, quite similar to the frequency found in Rio de Janeiro (4.4%) among European contributors." Silva et al. (2006)
- ↑ As of 11 November 2008 for example, the E-M35 phylogeny project[yes|permanent dead link|dead link}}] had records of four E-M123* tests, compared to 93 test results with E-M34.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Trombetta et al. 2015, Phylogeographic refinement and large scale genotyping of human Y chromosome haplogroup E provide new insights into the dispersal of early pastoralists in the African continent
- ↑ 2.0 2.1 "A Rare Deep-Rooting D0 African Y-chromosomal Haplogroup and its Implications for the Expansion of Modern Humans Out of Africa". Genetics 212 (4): 1421–1428. June 2019. doi:10.1534/genetics.119.302368. PMID 31196864.
- ↑ 3.0 3.1 3.2 3.3 Cruciani et al. (2007)
- ↑ ISOGG (2011)
- ↑ 5.0 5.1 5.2 Karafet et al. (2008)
- ↑ 6.0 6.1 6.2 Y Chromosome Consortium "YCC" (2002)
- ↑ Large-scale recent expansion of European patrilineages shown by population resequencing, Chiara Batini et al, nature.com, 2015
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 Cruciani et al. (2004)
- ↑ 9.0 9.1 Henn et al. (2008)
- ↑ Hassan et al. (2008)
- ↑ Lacan et al. (2011)
- ↑ "Molecular genetic investigation of the Neolithic population history in the western Carpathian Basin". http://ubm.opus.hbz-nrw.de/volltexte/2015/4075/pdf/doc.pdf.
- ↑ Schuenemann, Verena J. (2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications 8: 15694. doi:10.1038/ncomms15694. PMID 28556824. Bibcode: 2017NatCo...815694S.
- ↑ "Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe". 2018. bioRxiv 10.1101/191569.
- ↑ Rodrı́guez-Varela (2017). "Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modern North Africans". Current Biology 27 (1–7): 3396–3402.e5. doi:10.1016/j.cub.2017.09.059. PMID 29107554.
- ↑ Van De Loosdrecht, Marieke; Bouzouggar, Abdeljalil; Humphrey, Louise; Posth, Cosimo; Barton, Nick; Aximu-Petri, Ayinuer; Nickel, Birgit; Nagel, Sarah et al. (4 May 2018). "Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations". Science 360 (6388): 548–552. doi:10.1126/science.aar8380. PMID 29545507. Bibcode: 2018Sci...360..548V. https://www.science.org/doi/10.1126/science.aar8380.
- ↑ 17.0 17.1 17.2 17.3 Semino et al. (2004)
- ↑ Rosser et al. (2000)
- ↑ Firasat et al. (2006)
- ↑ Ehret, Keita & Newman (2004); Keita & Boyce (2005); Keita (2008).
- ↑ Behar et al. (2003)
- ↑ Behar et al. (2004)
- ↑ Shen et al. (2004)
- ↑ 24.0 24.1 Adams et al. (2008)
- ↑ Nebel et al. (2001)
- ↑ 26.0 26.1 26.2 Moran, Colin N. (2004). "Y chromosome haplogroups of elite Ethiopian endurance runners". Human Genetics 115 (6): 492–7. doi:10.1007/s00439-004-1202-y. PMID 15503146. https://www.researchgate.net/publication/8214295. Retrieved 6 February 2017.
- ↑ ISOGG (2008)
- ↑ 28.0 28.1 28.2 ISOGG 2015
- ↑ Cadenas et al. (2007)
- ↑ Trombetta et al. (2011)
- ↑ (Figure S.7) J D Cristofaro et al., 2013, "Afghan Hindu Kush: Where Eurasian Sub-Continent Gene Flows Converge", http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0076748
- ↑ 32.0 32.1 Cruciani et al. (2006)
- ↑ Peričic et al. (2005)
- ↑ "E-Y133414 YTree". https://yfull.com/tree/E-Y133414/.
- ↑ "E-FGC18960 YTree". https://yfull.com/tree/E-FGC18960*/.
- ↑ "E-L19 YTree". https://yfull.com/tree/E-L19/.
- ↑ "E-M81 YTree". https://www.yfull.com/tree/E-M81/.
- ↑ Cruciani et al. (2004).
- ↑ Fadhlaoui-Zid, Karima; Martinez-Cruz, Begoña; Khodjet-el-khil, Houssein; Mendizabal, Isabel; Benammar-Elgaaied, Amel; Comas, David (October 2011). "Genetic structure of Tunisian ethnic groups revealed by paternal lineages". American Journal of Physical Anthropology 146 (2): 271–280. doi:10.1002/ajpa.21581. ISSN 1096-8644. PMID 21915847.
- ↑ 40.0 40.1 Arredi et al. (2004)
- ↑ Alvarez et al. (2009)
- ↑ Bosch et al. (2001)
- ↑ Kujanová, Martina; Pereira, Luísa; Fernandes, Verónica; Pereira, Joana B.; Cerný, Viktor (October 2009). "Near eastern neolithic genetic input in a small oasis of the Egyptian Western Desert". American Journal of Physical Anthropology 140 (2): 336–346. doi:10.1002/ajpa.21078. ISSN 1096-8644. PMID 19425100.
- ↑ Robino et al. (2008)
- ↑ "E-A5604 YTree". https://yfull.com/tree/E-A5604/.
- ↑ Keita (2008)
- ↑ see table .
- ↑ Flores et al. (2005)
- ↑ Beleza et al. (2006)
- ↑ 50.0 50.1 Capelli et al. (2009)
- ↑ 51.0 51.1 Maca-Meyer et al. (2003)
- ↑ Fregel et al. (2009), see table
- ↑ Ramos-Luisa et al. (2009)
- ↑ Only men with French surname were analysed, in order to try to exclude more recent immigrants.
- ↑ Grugni, Viola; Raveane, Alessandro; Colombo, Giulia; Nici, Carmen; Crobu, Francesca; Ongaro, Linda; Battaglia, Vincenza; Sanna, Daria et al. (16 October 2019). "Y-chromosome and Surname Analyses for Reconstructing Past Population Structures: The Sardinian Population as a Test Case" (in en). International Journal of Molecular Sciences 20 (22): 5763. doi:10.3390/ijms20225763. ISSN 1422-0067. PMID 31744094.
- ↑ Francalacci et al. (2013), Low-Pass DNA Sequencing of 1200 Sardinians Reconstructs European Y-Chromosome Phylogeny
- ↑ 57.0 57.1 Di Gaetano et al. (2009)
- ↑ Sarno, S; Boattini, A; Carta, M; Ferri, G; Alù, M; Yao, DY; Ciani, G; Pettener, D et al. (2014). "An Ancient Mediterranean Melting Pot: Investigating the Uniparental Genetic Structure and Population History of Sicily and Southern Italy". PLOS ONE 9 (4): e96074. doi:10.1371/journal.pone.0096074. PMID 24788788. Bibcode: 2014PLoSO...996074S.
 This article contains quotations from this source, which is available under a Creative Commons Attribution 4.0 International (CC BY 4.0) license.
This article contains quotations from this source, which is available under a Creative Commons Attribution 4.0 International (CC BY 4.0) license.
- ↑ (8 out of 132), Mendizabal et al. (2008)
- ↑ (7 out of 295), Paracchini et al. (2003)
- ↑ Ordóñez, A. C.; Fregel, R.; Trujillo-Mederos, A.; Hervella, M.; de-la-Rúa, C.; Arnay-de-la-Rosa, M. (2017). "Genetic studies on the prehispanic population buried in Punta Azul cave (El Hierro, Canary Islands)". Journal of Archaeological Science 78: 20–28. doi:10.1016/j.jas.2016.11.004.
- ↑ "E-M81 YTree". https://yfull.com/tree/E-M81/.
- ↑ [1][yes|permanent dead link|dead link}}]
- ↑ "E-M35 Project Data". http://www.haplozone.net/e3b/project/cluster/3.
- ↑ "E-M35 Project Data". http://www.haplozone.net/e3b/project/cluster/4.
- ↑ "E-M35 Project Data". http://www.haplozone.net/e3b/project/cluster/81.
- ↑ "E-M35 Project Data". http://www.haplozone.net/e3b/project/cluster/72.
- ↑ "E-M35 Project Data". http://www.haplozone.net/e3b/project/cluster/60.
Additional sources
- Adams, Susan M; Bosch, Elena; Balaresque, Patricia L.; Ballereau, Stéphane J.; Lee, Andrew C.; Arroyo, Eduardo; López-Parra, Ana M.; Aler, Mercedes et al. (2008), "The Genetic Legacy of Religious Diversity and Intolerance: Paternal Lineages of Christians, Jews, and Muslims in the Iberian Peninsula", The American Journal of Human Genetics 83 (6): 725–36, doi:10.1016/j.ajhg.2008.11.007, PMID 19061982
- Alvarez; Santos, Cristina; Montiel, Rafael; Caeiro, Blazquez; Baali, Abdellatif; Dugoujon, Jean-Michel; Aluja, Maria Pilar (2009), "Y-chromosome variation in South Iberia: Insights into the North African contribution", American Journal of Human Biology 21 (3): 407–409, doi:10.1002/ajhb.20888, PMID 19213004
- Arredi, B; Poloni, E; Paracchini, S; Zerjal, T; Fathallah, D; Makrelouf, M; Pascali, V; Novelletto, A et al. (2004), "A Predominantly Neolithic Origin for Y-Chromosomal DNA Variation in North Africa", American Journal of Human Genetics 75 (2): 338–345, doi:10.1086/423147, PMID 15202071
- Badro, Danielle A.; Douaihy, Bouchra; Haber, Marc; Youhanna, Sonia C.; Salloum, Angélique; Ghassibe-Sabbagh, Michella; Johnsrud, Brian; Khazen, Georges et al. (2013), "Y-Chromosome and mtDNA Genetics Reveal Significant Contrasts in Affinities of Modern Middle Eastern Populations with European and African Populations", PLOS ONE 8 (1: e54616): e54616, doi:10.1371/journal.pone.0054616, PMID 23382925, Bibcode: 2013PLoSO...854616B
- Battaglia, Vincenza; Fornarino, Simona; Al-Zahery, Nadia; Olivieri, Anna; Pala, Maria; Myres, Natalie M; King, Roy J; Rootsi, Siiri et al. (2008), "Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe", European Journal of Human Genetics 17 (6): 820–830, doi:10.1038/ejhg.2008.249, PMID 19107149
- Behar, Doron M.; Thomas, Mark G.; Skorecki, Karl; Hammer, Michael F.; Bulygina, Ekaterina; Rosengarten, Dror; Jones, Abigail L.; Held, Karen et al. (October 2003), "Multiple Origins of Ashkenazi Levites: Y Chromosome Evidence for Both Near Eastern and European Ancestries", Am. J. Hum. Genet. 73 (4): 768–779, doi:10.1086/378506, PMID 13680527. Also at http://www.ucl.ac.uk/tcga/tcgapdf/Behar-AJHG-03.pdf and https://web.archive.org/web/20090304100321/http://www.familytreedna.com/pdf/400971.pdf
- Behar; Garrigan; Kaplan; Mobasher; Rosengarten (November 2004), "Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations", Hum. Genet. 114 (4): 354–365, doi:10.1007/s00439-003-1073-7, PMID 14740294, http://www.familytreedna.com/pdf/Behar_contrasting.pdf, retrieved 2009-09-11
- Beleza, Sandra; Gusmao, Leonor; Lopes, Alexandra; Alves, Cintia; Gomes, Iva; Giouzeli, Maria; Calafell, Francesc; Carracedo, Angel et al. (2006), "Micro-Phylogeographic and Demographic History of Portuguese Male Lineages", Annals of Human Genetics 70 (2): 181–194, doi:10.1111/j.1529-8817.2005.00221.x, PMID 16626329
- Bird, Steven (2007), "Haplogroup E3b1a2 as a Possible Indicator of Settlement in Roman Britain by Soldiers of Balkan Origin", Journal of Genetic Genealogy 3 (2), http://www.jogg.info/32/bird.htm
- Bortolini; Thomas, Mark G.; Chikhi, Lourdes; Aguilar, Juan A.; Castro-De-Guerra, Dinorah; Salzano, Francisco M.; Ruiz-Linares, Andres (2004), "Ribeiro's typology, genomes, and Spanish colonialism, as viewed from Gran Canaria and Colombia", Genetics and Molecular Biology 27 (1): 1–8, doi:10.1590/S1415-47572004000100001, http://www.scielo.br/pdf/gmb/v27n1/a01v27n1.pdf
- Bosch, Elena; Calafell, Francesc; Comas, David; Oefner, Peter J.; Underhill, Peter A.; Bertranpetit, Jaume (2001), "High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between north-western Africa and the Iberian Peninsula", Am J Hum Genet 68 (4): 1019–1029, doi:10.1086/319521, PMID 11254456
- Bosch, E.; Calafell, F.; Gonzalez-Neira, A.; Flaiz, C.; Mateu, E.; Scheil, H.-G.; Huckenbeck, W.; Efremovska, L. et al. (2006), "Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns", Annals of Human Genetics 70 (4): 459–487, doi:10.1111/j.1469-1809.2005.00251.x, PMID 16759179, http://www3.interscience.wiley.com/journal/118548826/abstract?CRETRY=1&SRETRY=0
- Cadenas; Zhivotovsky, Lev A; Cavalli-Sforza, Luca L; Underhill, Peter A; Herrera, Rene J (2007), "Y-chromosome diversity characterizes the Gulf of Oman", European Journal of Human Genetics 16 (3): 1–13, doi:10.1038/sj.ejhg.5201934, PMID 17928816
- Capelli, Cristian; Redhead, Nicola; Abernethy, Julia K.; Gratrix, Fiona; Wilson, James F.; Moen, Torolf; Hervig, Tor; Richards, Martin et al. (2003), "A Y Chromosome Census of the British Isles", Current Biology 13 (11): 979–84, doi:10.1016/S0960-9822(03)00373-7, PMID 12781138 also at [2]
- Caratti; Gino, S.; Torre, C.; Robino, C. (2009), "Subtyping of Y-chromosomal haplogroup E-M78 (E1b1b1a) by SNP assay and its forensic application", International Journal of Legal Medicine 123 (4): 357–360, doi:10.1007/s00414-009-0350-y, PMID 19430804
- Capelli, Cristian; Onofri, Valerio; Brisighelli, Francesca; Boschi, Ilaria; Scarnicci, Francesca; Masullo, Mara; Ferri, Gianmarco; Tofanelli, Sergio et al. (2009), "Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe", European Journal of Human Genetics 17 (6): 848–852, doi:10.1038/ejhg.2008.258, PMID 19156170
- Cinnioğlu, Cengiz; King, Roy; Kivisild, Toomas; Kalfoglu, Ersi; Atasoy, Sevil; Cavalleri, Gianpiero L.; Lillie, Anita S.; Roseman, Charles C. et al. (2004), "Excavating Y-chromosome haplotype strata in Anatolia", Hum Genet 114 (2): 127–48, doi:10.1007/s00439-003-1031-4, PMID 14586639
- Contu, Daniela; Morelli, Daniela; Santoni, Federico; Foster, Jamie W.; Francalacci, Paolo; Cucca, Francesco (2008), "Y-Chromosome Based Evidence for Pre-Neolithic Origin of the Genetically Homogeneous but Diverse Sardinian Population: Inference for Association Scans", PLOS ONE 3 (1): e1430, doi:10.1371/journal.pone.0001430, PMID 18183308, Bibcode: 2008PLoSO...3.1430C
- Cruciani, Fulvio; Santolamazza, Piero; Shen, Peidong; MacAulay, Vincent; Moral, Pedro; Olckers, Antonel; Modiano, David; Holmes, Susan (2002), "A Back Migration from Asia to Sub-Saharan Africa Is Supported by High-Resolution Analysis of Human Y-Chromosome Haplotypes", American Journal of Human Genetics 70 (5): 1197–1214, doi:10.1086/340257, PMID 11910562
- Cruciani; La Fratta; Santolamazza; Sellitto (May 2004), "Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out Of Africa", American Journal of Human Genetics 74 (5): 1014–1022, doi:10.1086/386294, PMID 15042509, PMC 1181964, http://www.familytreedna.com/pdf/hape3b.pdf, retrieved 2008-05-17
- Cruciani; La Fratta; Torroni; Underhill; Scozzari (2006), "Molecular Dissection of the Y Chromosome Haplogroup E-M78 (E3b1a): A Posteriori Evaluation of a Microsatellite-Network-Based Approach Through Six New Biallelic Markers", Human Mutation 27 (8): 831–2, doi:10.1002/humu.9445, PMID 16835895
- Cruciani, F.; La Fratta, R.; Trombetta, B.; Santolamazza, P.; Sellitto, D.; Colomb, E. B.; Dugoujon, J.-M.; Crivellaro, F. et al. (2007), "Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12", Molecular Biology and Evolution 24 (6): 1300–1311, doi:10.1093/molbev/msm049, PMID 17351267, http://mbe.oxfordjournals.org/cgi/reprint/24/6/1300 Also see Supplementary Data.
- Di Gaetano; Cerutti, Francesca; Crobu, Carlo; Robino (2009), "Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome", European Journal of Human Genetics 17 (1): 91–99, doi:10.1038/ejhg.2008.120, PMID 18685561
- Dugoujon; Coudray; Torroni; Cruciani; Scozzari; Moral; Louali; Kossmann (2009), d'Errico; Hombert, eds., "The Berber and the Berbers: Genetic and linguistic diversities", Becoming Eloquent Advances in the Emergence of Language, Human Cognition, and Modern Cultures: 123–146, doi:10.1075/z.152.05ch4, ISBN 978-90-272-3269-4, http://npu.edu.ua/!e-book/book/djvu/A/iif_kgpm_Errico_Becoming_Eloquent_pdf.pdf
- Ehret, C.; Keita, SO; Newman, P (2004), "The Origins of Afroasiatic", Science 306 (5702): 1680, doi:10.1126/science.306.5702.1680c, PMID 15576591
- El-Sibai, Mirvat; Platt, Daniel E.; Haber, Marc; Xue, Yali; Youhanna, Sonia C.; Wells, R. Spencer; Izaabel, Hassan; Sanyoura, May F. et al. (2009), "Geographical Structure of the Y-chromosomal Genetic Landscape of the Levant: A coastal-inland contrast", Annals of Human Genetics 73 (6): 568–581, doi:10.1111/j.1469-1809.2009.00538.x, PMID 19686289
- Firasat; Khaliq, Shagufta; Mohyuddin, Aisha; Papaioannou, Myrto; Tyler-Smith, Chris; Underhill, Peter A; Ayub, Qasim (2006), "Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan", European Journal of Human Genetics 15 (1): 121–126, doi:10.1038/sj.ejhg.5201726, PMID 17047675
- Flores, Carlos; Maca-Meyer, Nicole; González, Ana M; Oefner, Peter J; Shen, Peidong; Pérez, Jose A; Rojas, Antonio; Larruga, Jose M et al. (2004), "Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography", European Journal of Human Genetics 12 (10): 855–863, doi:10.1038/sj.ejhg.5201225, PMID 15280900
- Flores; Maca-Meyer, Nicole; Larruga, Jose M.; Cabrera, Vicente M.; Karadsheh, Naif; Gonzalez, Ana M. (2005), "Isolates in a corridor of migrations: a high-resolution analysis of Y-chromosome variation in Jordan", J Hum Genet 50 (9): 435–441, doi:10.1007/s10038-005-0274-4, PMID 16142507
- Francalacci, P.; Morelli, L.; Underhill, P.A.; Lillie, A.S.; Passarino, G.; Useli, A.; Madeddu, R.; Paoli, G. et al. (2003), "Peopling of Three Mediterranean Islands (Corsica, Sardinia, and Sicily) Inferred by Y-Chromosome Biallelic Variability", American Journal of Physical Anthropology 121 (3): 270–279, doi:10.1002/ajpa.10265, PMID 12772214
- Fregel, Rosa; Gomes, Verónica; Gusmão, Leonor; González, Ana M; Cabrera, Vicente M; Amorim, António; Larruga, Jose M (2009), "Demographic history of Canary Islands male gene-pool: replacement of native lineages by European", BMC Evolutionary Biology 9: 181, doi:10.1186/1471-2148-9-181, PMID 19650893
- Gérard; Berriche, S; Aouizérate, A; Diéterlen, F; Lucotte, G (2006), "North African Berber and Arab influences in the western Mediterranean revealed by Y-chromosome DNA haplotypes", Human Biology 78 (3): 307–316, doi:10.1353/hub.2006.0045, PMID 17216803, http://cat.inist.fr/?aModele=afficheN&cpsidt=18328663
- Hammer (2003), "Human population structure and its effects on sampling Y chromosome sequence variation", Genetics 164 (4): 1495–1509, doi:10.1093/genetics/164.4.1495, PMID 12930755
- Hassan, Hisham Y.; Underhill, Peter A.; Cavalli-Sforza, Luca L.; Ibrahim, Muntaser E. (2008), "Y-Chromosome Variation Among Sudanese: Restricted Gene Flow, Concordance With Language, Geography, and History", American Journal of Physical Anthropology 137 (3): 316–23, doi:10.1002/ajpa.20876, PMID 18618658, http://dirkschweitzer.net/E3b-papers/Hassan-Sudan-2008-AJPA.pdf
- Henn, B. M.; Gignoux, C.; Lin, Alice A; Oefner, Peter J.; Shen, P.; Scozzari, R.; Cruciani, F.; Tishkoff, S. A. et al. (2008), "Y-chromosomal evidence of a pastoralist migration through Tanzania to southern Africa", PNAS 105 (31): 10693–8, doi:10.1073/pnas.0801184105, PMID 18678889, Bibcode: 2008PNAS..10510693H. See comment on Dienekes blog, comment on the Spitoon blog and public release.
- Jobling, M.A.; Tyler-Smith, C. (2000), "New uses for new haplotypes the human Y chromosome, disease and selection", Trends Genet. 16 (8): 356–362, doi:10.1016/S0168-9525(00)02057-6, PMID 10904265
- Karafet, T. M.; Mendez, F. L.; Meilerman, M. B.; Underhill, P. A.; Zegura, S. L.; Hammer, M. F. (May 2008), "New Binary Polymorphisms Reshape and Increase Resolution of the Human Y-Chromosomal Haplogroup Tree", Genome Research 18 (5): 830–8, doi:10.1101/gr.7172008, PMID 18385274, PMC 2336805, http://www.genome.org/cgi/content/abstract/gr.7172008v1. Published online April 2, 2008. See also Supplementary Material.
- Keita, Shomarka (2008), "Geography, selected Afro-Asiatic families, and Y chromosome lineage variation", In Hot Pursuit of Language in Prehistory: Essays in the Four Fields of Anthropology : In Honor of Harold Crane Fleming, ISBN 978-90-272-3252-6, https://books.google.com/books?id=xxcdjUGfx40C&pg=PA3
- Keita, S. O. Y.; Boyce, A. J. (Anthony J.) (2005), "Genetics, Egypt, and History: Interpreting Geographical Patterns of Y Chromosome Variation", History in Africa 32: 221–246, doi:10.1353/hia.2005.0013, http://muse.jhu.edu/journals/history_in_africa/v032/32.1keita.html
- King, R. J.; Özcan, S. S.; Carter, T.; Kalfoğlu, E.; Atasoy, S.; Triantaphyllidis, C.; Kouvatsi, A.; Lin, A. A. et al. (2008), "Differential Y-chromosome Anatolian Influences on the Greek and Cretan Neolithic", Annals of Human Genetics 72 (2): 205–214, doi:10.1111/j.1469-1809.2007.00414.x, PMID 18269686, http://dirkschweitzer.net/E3b-papers/KingAHG-08-72-205.pdf
- King; Underhill (2002), "Congruent distribution of Neolithic painted pottery and ceramic figurines with Y-chromosome lineages", Antiquity 76 (293): 707–14, doi:10.1017/S0003598X00091158
- Kujanova; Pereira; Fernandes; Pereira; Cerný (2009), "Near Eastern Neolithic Genetic Input in a Small Oasis of the Egyptian Western Desert", American Journal of Physical Anthropology 140 (2): 336–346, doi:10.1002/ajpa.21078, PMID 19425100
- Lacan, Marie; Keyser, Christine; Ricaut, François-Xavier; Brucato, Nicolas; Tarrús, Josep; Bosch, Angel; Guilaine, Jean; Crubézy, Eric et al. (2011), "Ancient DNA suggests the leading role played by men in the Neolithic dissemination", PNAS 108 (45): 18255–9, doi:10.1073/pnas.1113061108, PMID 22042855, Bibcode: 2011PNAS..10818255L
- Lancaster, Andrew (2009), "Y Haplogroups, Archaeological Cultures and Language Families: a Review of the Multidisciplinary Comparisons using the case of E-M35", Journal of Genetic Genealogy 5 (1), http://www.jogg.info/51/files/Lancaster.pdf
- Luis, J; Rowold, D; Regueiro, M; Caeiro, B; Cinnioglu, C; Roseman, C; Underhill, P; Cavallisforza, L et al. (2004), "The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations", American Journal of Human Genetics 74 (3): 532–544, doi:10.1086/382286, PMID 14973781, PMC 1182266, http://hpgl.stanford.edu/publications/AJHG_2004_v74_p000-0130.pdf. (Also see Errata)
- Maca-Meyer, N. et al. (2003), "Y Chromosome and Mitochondrial DNA Characterization of Pasiegos, a Human Isolate from Cantabria (Spain)", Annals of Human Genetics 67 (Pt 4): 329–339, doi:10.1046/j.1469-1809.2003.00045.x, PMID 12914567.
- Martinez, Laisel; Underhill, Peter A; Zhivotovsky, Lev A; Gayden, Tenzin; Moschonas, Nicholas K; Chow, Cheryl-Emiliane T; Conti, Simon; Mamolini, Elisabetta et al. (April 1, 2007), "Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau", European Journal of Human Genetics 15 (4): 485–493, doi:10.1038/sj.ejhg.5201769, ISSN 1018-4813, PMID 17264870
- Mendizabal, Isabel; Sandoval, Karla; Berniell-Lee, Gemma; Calafell, Francesc; Salas, Antonio; Martinez-Fuentes, Antonio; Comas, David (2008), "Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba", BMC Evol. Biol. 8: 213, doi:10.1186/1471-2148-8-213, PMID 18644108
- Nebel, Almut; Filon, D; Brinkmann, B; Majumder, P; Faerman, M; Oppenheim, A (2001), "The Y Chromosome Pool of Jews as Part of the Genetic Landscape of the Middle East", American Journal of Human Genetics 69 (5): 1095–1112, doi:10.1086/324070, PMID 11573163
- Paracchini; Pearce, CL; Kolonel, LN; Altshuler, D; Henderson, BE; Tyler-Smith, C (2003), "A Y chromosomal influence on prostate cancer risk: the multi-ethnic cohort study", J Med Genet 40 (11): 815–819, doi:10.1136/jmg.40.11.815, PMID 14627670
- Pelotti; Ceccardi, S; Lugaresi, F; Trane, R; Falconi, M; Bini, C; Willuweit, S; Roewer, L (2007), "Microgeographic genetic variation of Y chromosome in a population sample of Ravenna's area in the Emilia-Romagna region (North of Italy)", Forensic Science International: Genetics Supplement Series 1 (1): 242–243, doi:10.1016/j.fsigss.2007.10.025
- Pereira, Luísa; Černý, Viktor; Cerezo, María; Silva, Nuno M; Hájek, Martin; Vašíková, Alžběta; Kujanová, Martina; Brdička, Radim et al. (2010), "Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel", European Journal of Human Genetics 18 (8): 915–923, doi:10.1038/ejhg.2010.21, PMID 20234393, PMC 2987384, http://peer.ccsd.cnrs.fr/docs/00/51/83/10/PDF/PEER_stage2_10.1038%252Fejhg.2010.21.pdf
- Peričic, M.; Lauc, LB; Klarić, IM; Rootsi, S; Janićijevic, B; Rudan, I; Terzić, R; Colak, I et al. (2005), "High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations", Mol. Biol. Evol. 22 (10): 1964–75, doi:10.1093/molbev/msi185, PMID 15944443, http://mbe.oxfordjournals.org/cgi/content/full/22/10/1964.
- Pontikos D. "Phylogeographic refinement of haplogroup E" http://dienekes.blogspot.ru/2015/07/phylogeographic-refinement-of.html
- Regueiro, M.; Cadenas, A.M.; Gayden, T.; Underhill, P.A.; Herrera, R.J. (2006), "Iran: Tricontinental Nexus for Y-Chromosome Driven Migration", Hum Hered 61 (3): 132–143, doi:10.1159/000093774, PMID 16770078
- Robino, C.; Crobu, F.; Gaetano, C.; Bekada, A.; Benhamamouch, S.; Cerutti, N.; Piazza, A.; Inturri, S. et al. (2008), "Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample", Journal International Journal of Legal Medicine 122 (3): 251–5, doi:10.1007/s00414-007-0203-5, PMID 17909833
- Rosa, Alexandra; Ornelas, Carolina; Jobling, Mark A; Brehm, António; Villems, Richard (2007), "Y-chromosomal diversity in the population of Guinea-Bissau: a multiethnic perspective", BMC Evolutionary Biology 7: 124, doi:10.1186/1471-2148-7-124, PMID 17662131
- Rosser, Z; Zerjal, T; Hurles, M; Adojaan, M; Alavantic, D; Amorim, A; Amos, W; Armenteros, M et al. (2000), "Y-Chromosomal Diversity in Europe Is Clinal and Influenced Primarily by Geography, Rather than by Language", American Journal of Human Genetics 67 (6): 1526–1543, doi:10.1086/316890, PMID 11078479, PMC 1287948, http://www.ajhg.org/AJHG/abstract/S0002-9297(07)63221-2
- Sanchez, Juan J; Hallenberg, Charlotte; Børsting, Claus; Hernandez, Alexis; Gorlin, RJ (2005), "High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males", European Journal of Human Genetics 13 (7): 856–866, doi:10.1038/sj.ejhg.5201390, PMID 15756297. Published online 9 March 2005
- Scozzari, Rosaria; Cruciani, F; Pangrazio, A; Santolamazza, P; Vona, G; Moral, P; Latini, V; Varesi, L et al. (2001), "Human Y-Chromosome Variation in the Western Mediterranean Area: Implications for the Peopling of the Region", Human Immunology 62 (9): 871–884, doi:10.1016/S0198-8859(01)00286-5, PMID 11543889, http://evolutsioon.ut.ee/publications/Scozzari2001.pdf
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA; Arbuzova, S; Beckman, LE; De Benedictis, G; Francalacci, P et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science 290 (5494): 1155–59, doi:10.1126/science.290.5494.1155, PMID 11073453, Bibcode: 2000Sci...290.1155S, http://hpgl.stanford.edu/publications/Science_2000_v290_p1155.pdf.
- Semino; Santachiara-Benerecetti, A. Silvana; Falaschi, Francesco; Cavalli-Sforza, L. Luca; Underhill, Peter A. (2002), "Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny", Am J Hum Genet 70 (1): 265–268, doi:10.1086/338306, PMID 11719903, PMC 384897, http://hpgl.stanford.edu/publications/AJHG_2002_v70_p265-268.pdf
- Semino, Ornella; Magri, Chiara; Benuzzi, Giorgia; Lin, Alice A.; Al-Zahery, Nadia; Battaglia, Vincenza; MacCioni, Liliana; Triantaphyllidis, Costas et al. (2004), "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area", American Journal of Human Genetics 74 (5): 1023–1034, doi:10.1086/386295, PMID 15069642
- Shen, Peidong; Lavi, Tal; Kivisild, Toomas; Chou, Vivian; Sengun, Deniz; Gefel, Dov; Shpirer, Issac; Woolf, Eilon et al. (2004), "Reconstruction of Patrilineages and Matrilineages of Samaritans and Other Israeli Populations From Y-Chromosome and Mitochondrial DNA Sequence Variation", Human Mutation 24 (3): 248–60, doi:10.1002/humu.20077, PMID 15300852, http://www.ebc.ee/EVOLUTSIOON/publications/Shen2004.pdf, retrieved 2008-11-07
- Silva; Carvalho, Elizeu; Costa, Guilherme; Tavares, Lígia; Amorim, António; Gusmão, Leonor (2006), "Y-chromosome genetic variation in Rio de Janeiro population", American Journal of Human Biology 18 (6): 829–837, doi:10.1002/ajhb.20567, PMID 17039481, http://www3.interscience.wiley.com/journal/113395738/abstract?CRETRY=1&SRETRY=0
- Shlush; Behar, Doron M.; Yudkovsky, Guennady; Templeton, Alan; Hadid, Yarin; Basis, Fuad; Hammer, Michael; Itzkovitz, Shalev et al. (2008), Gemmell, Neil John, ed., "The Druze: A Population Genetic Refugium of the Near East", PLOS ONE 3 (5): e2105, doi:10.1371/journal.pone.0002105, PMID 18461126, Bibcode: 2008PLoSO...3.2105S
- Solé-Morata, Neus; García-Fernández, Carla; Urasin, Vadim; Bekada, Asmahan; Fadhlaoui-Zid, Karima; Zalloua, Pierre; Comas, David; Calafell, Francesc (21 November 2017), "Whole Y-chromosome sequences reveal an extremely recent origin of the most common North African paternal lineage E-M183 (M81)" (in en), Scientific Reports 7 (1): 15941, doi:10.1038/s41598-017-16271-y, ISSN 2045-2322, PMID 29162904, Bibcode: 2017NatSR...715941S
- Sykes, Bryan (2006), Blood of the Isles: Exploring the Genetic Roots of Our Tribal History, Bantam, ISBN 978-0-593-05652-3, https://books.google.com/books?id=tJkyAAAACAAJ&q=%22Blood+of+the+Isles%22
- Tishkoff; Gonder; Henn; Mortensen; Knight (2007), "History of Click-Speaking Populations of Africa Inferred from mtDNA and Y Chromosome Genetic Variation", Mol Biol Evol 24 (10): 2180–2195, doi:10.1093/molbev/msm155, PMID 17656633, http://mbe.oxfordjournals.org/content/24/10/2180.full
- Thomas; Stumpf, M. P.H; Harke, H. (2006), "Evidence for an apartheid-like social structure in early Anglo-Saxon England", Proceedings of the Royal Society B 273 (1601): 2651–2657, doi:10.1098/rspb.2006.3627, PMID 17002951, PMC 1635457, http://publishing.royalsociety.org/media/proceedings_b/papers/RSPB20063627.pdf
- Trombetta, Beniamino; Cruciani, Fulvio; Sellitto, Daniele; Scozzari, Rosaria (2011), MacAulay, Vincent, ed., "A New Topology of the Human Y Chromosome Haplogroup E1b1 (E-P2) Revealed through the Use of Newly Characterized Binary Polymorphisms", PLOS ONE 6 (1): e16073, doi:10.1371/journal.pone.0016073, PMID 21253605, Bibcode: 2011PLoSO...616073T
- Trombetta B. "Phylogeographic Refinement and Large Scale Genotyping of Human Y Chromosome Haplogroup E Provide New Insights into the Dispersal of Early Pastoralists in the African Continent" http://gbe.oxfordjournals.org/content/7/7/1940.long
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li; Passarino, Giuseppe; Yang, Wei H.; Kauffman, Erin; Bonné-Tamir, Batsheva et al. (2000), "Y chromosome sequence variation and the history of human populations", Nat Genet 26 (3): 358–361, doi:10.1038/81685, PMID 11062480
- Underhill; Passarino, G.; Lin, A. A.; Shen, P.; Mirazon Lahr, M.; Foley, R. A.; Oefner, P. J.; Cavalli-Sforza, L. L. (2001), "The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations", Ann Hum Genet 65 (Pt 1): 43–62, doi:10.1046/j.1469-1809.2001.6510043.x, PMID 11415522
- Underhill (2002), "Inference of Neolithic Population Histories using Y-chromosome Haplotypes", in Bellwood, Peter; Renfrew, A. Colin, Examining the farming/language dispersal hypothesis, Cambridge: McDonald Institute for Archaeological Research, ISBN 978-1-902937-20-5
- Underhill, Peter A.; Kivisild, Toomas (2007), "Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations", Annu. Rev. Genet. 41: 539–64, doi:10.1146/annurev.genet.41.110306.130407, PMID 18076332
- Weale, M. E.; Weiss, D. A.; Jager, R. F.; Bradman, N.; Thomas, M. G. (2002), "Y Chromosome Evidence for Anglo-Saxon Mass Migration", Mol. Biol. Evol. 19 (7): 1008–1021, doi:10.1093/oxfordjournals.molbev.a004160, PMID 12082121, http://mbe.oxfordjournals.org/cgi/reprint/19/7/1008.pdf.
- Weale; Shah, T; Jones, AL; Greenhalgh, J; Wilson, JF; Nymadawa, P; Zeitlin, D; Connell, BA et al. (September 1, 2003), "Rare Deep-Rooting Y Chromosome Lineages in Humans: Lessons for Phylogeography", Genetics 165 (1): 229–234, doi:10.1093/genetics/165.1.229, PMID 14504230, PMC 1462739, http://www.genetics.org/cgi/reprint/165/1/229
- Wood; Stover; Ehret; Destro-Bisol (2005), "Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes", European Journal of Human Genetics 13 (7): 867–876, doi:10.1038/sj.ejhg.5201408, PMID 15856073
- Y Chromosome Consortium "YCC" (2002), "A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups", Genome Research 12 (2): 339–348, doi:10.1101/gr.217602, PMID 11827954
- Zalloua, Pierre A.; Xue, Yali; Khalife, Jade; Makhoul, Nadine; Debiane, Labib; Platt, Daniel E.; Royyuru, Ajay K.; Herrera, Rene J. et al. (2008), "Y-Chromosomal Diversity in Lebanon Is Structured by Recent Historical Events", American Journal of Human Genetics 82 (4): 873–882, doi:10.1016/j.ajhg.2008.01.020, PMID 18374297
- Zalloua, Pierre A.; Platt, Daniel E.; El Sibai, Mirvat; Khalife, Jade; Makhoul, Nadine; Haber, Marc; Xue, Yali; Izaabel, Hassan et al. (2008), "Identifying Genetic Traces of Historical Expansions: Phoenician Footprints in the Mediterranean", The American Journal of Human Genetics 83 (5): 633–642, doi:10.1016/j.ajhg.2008.10.012, PMID 18976729
- Zerjal (1999), The use of Y-chromosomal DNA variation to investigate population history; in Papiha SS, Deka R, Chakraborty R (eds): Genomic diversity: applications in human population genetics, Kluwer Academic/Plenum Publishers, pp. 91–101
- Zhao; Khan, Faisal; Borkar, Minal; Herrera, Rene; Agrawal, Suraksha (2009), "Presence of three different paternal lineages among North Indians: A study of 560 Y chromosomes", Annals of Human Biology 36 (1): 1–14, doi:10.1080/03014460802558522, PMID 19058044
Sources for conversion tables
- Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H. et al. (February 2001). "A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania". The American Journal of Human Genetics 68 (2): 432–443. doi:10.1086/318205. PMID 11170891.
- Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi et al. (1 July 2001). "Hierarchical Patterns of Global Human Y-Chromosome Diversity". Molecular Biology and Evolution 18 (7): 1189–1203. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora et al. (February 2001). "Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages". European Journal of Human Genetics 9 (2): 97–104. doi:10.1038/sj.ejhg.5200597. PMID 11313742.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William et al. (September 2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics 69 (3): 615–628. doi:10.1086/323299. PMID 11481588.
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science 290 (5494): 1155–9, doi:10.1126/science.290.5494.1155, PMID 11073453, Bibcode: 2000Sci...290.1155S
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics 65 (6): 1718–1724. doi:10.1086/302680. PMID 10577926.
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–361. doi:10.1038/81685. PMID 11062480.
External links
- E-M35 Phylogeny Project Wiki
- E-M35 Phylogeny Project (all labs site)
- E-M35 Phylogeny Project Public Forum
- Jewish E3b Project at FTDNA
- Map of E-M35 distribution in Europe
- Haplogroup E3B1 (E1b1b1) Facebook Group (M35 community)
- E1b1b1b*-A Project
- E1b1.org – International Y-DNA project of Haplogroup E1b1 and its Subclades

