Biology:Histocompatibility
Histocompatibility, or tissue compatibility, is the property of having the same, or sufficiently similar, alleles of a set of genes called human leukocyte antigens (HLA), or major histocompatibility complex (MHC).[1] Each individual expresses many unique HLA proteins on the surface of their cells, which signal to the immune system whether a cell is part of the self or an invading organism.[2] T cells recognize foreign HLA molecules and trigger an immune response to destroy the foreign cells.[3] Histocompatibility testing is most relevant for topics related to whole organ, tissue, or stem cell transplants, where the similarity or difference between the donor's HLA alleles and the recipient's triggers the immune system to reject the transplant.[4] The wide variety of potential HLA alleles lead to unique combinations in individuals and make matching difficult.
Discovery
The discovery of the MHC and role of histocompatibility in transplantation was a combined effort of many scientists in the 20th century. A genetic basis for transplantation rejection was proposed in a 1914 Nature paper by C.C. Little and Ernest Tyyzer, which showed that tumors transplanted between genetically identical mice grew normally, but tumors transplanted between non-identical mice were rejected and failed to grow.[5] The role of the immune system in transplant reject was proposed by Peter Medawar, whose skin graft transplants in world war two victims showed that skin transplants between individuals had much higher rejection rates than self-transplants within an individual, and that suppressing the immune system delayed skin transplant rejection.[6] Medawar shared the 1960 Nobel Prize in part for this work.[7]
In the 1930s and 1940s, George Snell and Peter Gorer individually isolated the genetic factors that when similar allowed transplantation between mouse strains, naming them H and antigen II respectively. These factors were in fact one and the same, and the locus was named H-2. Snell coined the term "histocompatibility" to describe the relationship between the H-2 cell-surface proteins and transplant acceptance.[8] The human version of the histocompatibility complex was found by Jean Dausset in the 1950s, when he noticed that recipients of blood transfusions were producing antibodies directed against only the donor cells.[9] The target of these antibodies, or the human leukocyte antigens (HLA), were discovered to be the human homologue of Snell and Gorer's mouse MHC. Snell, Dausset and Baruj Benacerraf shared the 1980 Nobel Prize for the discovery of the MHC and HLA.[10]
Major histocompatibility complex (MHC)
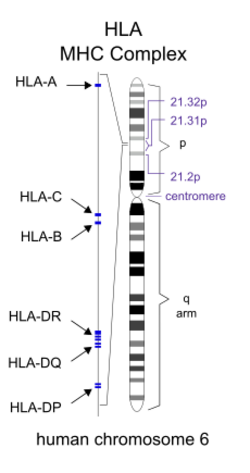
HLA, the human form of the major histocompatibility complex (MHC), is located on chromosome 6 at 6p21.3.[11] Individuals inherit two different HLA haplotypes, one from each parent, each containing more than 200 genes relevant to helping the immune system recognize foreign invaders. These genes include MHC class I and class II cell-surface proteins.[12] MHC Class I molecules—HLA-A, HLA-B, and HLA-C—are present on all nucleated cells and are responsible for signaling to an immune cell that an antigen is inside the cell.[2] MHC Class II molecules—HLA-DR, and HLA-DQ and HLA-DP—are only present on antigen presenting cells and are responsible for presenting molecules from invading organisms to cells of the immune system.[13]
The MHC genes are highly polymorphic, with thousands of versions of the MHC receptors in the population, though any one individual can have no more than two versions for any one locus.[14] MHC receptors are codominantly expressed, meaning all inherited alleles are expressed by the individual.[15] The wide variety of potential alleles and multiple loci in the HLA allow for many unique combinations in individuals.
Role in transplantation
After receiving a transplant, the recipient's T cells will become activated by foreign MHC molecules on the donor tissue and trigger the immune system to attack the donated tissue[3] The more similar HLA alleles are between donor and recipient, the fewer foreign targets exist on the donor tissue for the host immune system to recognize and attack.[16] The number and selection of MHC molecules to be considered when determining whether two individuals are histocompatible fluctuates based on application, however matching HLA-A, HLA-B, and HLA-DR has been shown to improve patient outcomes.[17] Histocompatibility has a measurable effect on whole organ transplantation, increasing life expectancy of both the patient and organ.[3] HLA similarity is therefore a relevant factor when choosing donors for tissue or organ transplant. This is especially important for pancreas and kidney transplants.
Due to the inherited nature of HLA genes, family members are more likely to be histocompatible. The odds of a sibling having received the same haplotypes from both parents is 25%, while there is a 50% chance that the sibling would share just one haplotype and a 25% chance they would share neither. However, variability due to crossing over, haplotypes may rearrange between generations and siblings may be intermediate matches.[18]
The degree of histocompatibility required is dependent on individual factors, including the type of tissue or organ and the medical condition of the recipient. While whole organ transplants can be successful between unmatched individuals, increased histocompatibility lowers rates of rejection, result in longer lifespans, and overall lower associated hospital costs.[19] The impact of HLA matching differs even among whole organ transplants, with some studies reporting less importance in liver transplants as compared to heart, lung, and other organs.[17] In comparison, hematopoietic stem cell transplants are often require higher degrees of matching due to the increased risk of graft-versus-host disease, in which the donor's immune system recognizes the recipient's MHC molecules as foreign and mounts an immune response.[20] Some transplanted tissue is not exposed to T cells that could detect foreign MHC molecules, such as corneas, and thus histocompatibility is not a factor in transplantation.[21] Individual factors such as age sometimes factors into matching protocol, as the immune response of older transplant patients towards MHC proteins is slower and therefore less compatibility is necessary for positive results.[22] Post-operative immunosuppressant therapy is often used to lessen the immune response and prevent tissue rejection by dampening the immune system's response to the foreign HLA molecules,[23] and can increase the likelihood of successful transplantation in non-identical transplant recipients.[24]
Testing
Because of the clinical significance of histocompatibility in tissue transplants, several methods of typing are used to check for HLA allele expression.
Serological Typing
Serological typing involves incubating lymphocytes from the recipient with serum containing known antibodies against the varying HLA alleles. If the serum contains an antibody specific for a HLA allele that is present on the recipient's lymphocyte, the antibodies will bind to the cell and activate a complement signaling cascade resulting in cell lysis. A lysed cell will take up an added dye such as trypan blue allowing for identification. Comparing which serums triggers cell lysis allows identification of HLA alleles present on the cell surface of the recipients cells.[25]
Serological typing has the benefit of quickly identifying expressed HLA alleles, and ignores any non-expressed alleles that could be of little immunological significance. However, it does not recognize subclasses of alleles, which are sometimes necessary for matching.[25]
Molecular Typing
HLA alleles can be determined by directly analyzing the HLA loci on chromosome 6. Sequence specific oligonucleotide probes, sequence specific primer PCR amplification, and direct sequencing can all be used to identify HLA alleles, often providing amino acid level resolution. Molecular methods can more accurately identify rare and unique alleles, but do not provide information about expression levels.[25]
See also
- Clonal selection theory
- Major histocompatibility complex (MHC)
- Tissue typing
- Transplant rejection
References
- ↑ "Histocompatibility". Dorlands Illustrated Medical Dictionary. Philadelphia, PA: Elsevier. 2012.
- ↑ 2.0 2.1 "Human leukocyte antigens". Genetics Home Reference. https://ghr.nlm.nih.gov/primer/genefamily/hla.
- ↑ 3.0 3.1 3.2 "Mechanism of cellular rejection in transplantation". Pediatric Nephrology 25 (1): 61–74. January 2010. doi:10.1007/s00467-008-1020-x. PMID 21476231.
- ↑ Kindt, Thomas J.; Goldsby, Richard A.; Osborne, Barbara Anne; Kuby, Janis (2006). Kurby Immunology. W. H. Freeman & Company. ISBN 978-1-4292-0211-4.
- ↑ "Clarence Cook Little (1888-1971): the genetic basis of transplant immunology". American Journal of Transplantation 4 (2): 155–9. February 2004. doi:10.1046/j.1600-6143.2003.00324.x. PMID 14974934.
- ↑ "Peter Brian Medawar: father of transplantation". Journal of the American College of Surgeons 180 (3): 332–6. March 1995. PMID 7874344.
- ↑ "The Nobel Prize in Physiology or Medicine 1960 - Speed Read". https://www.nobelprize.org/nobel_prizes/medicine/laureates/1960/speedread.html.
- ↑ Elgert, Klaus D. (2009). Immunology : understanding the immune system (2nd ed.). Hoboken, N.J.: Wiley-Blackwell. ISBN 9780470081570. OCLC 320494512.
- ↑ "The Nobel Prize in Physiology or Medicine 1980 - Speed Read". https://www.nobelprize.org/nobel_prizes/medicine/laureates/1980/speedread.html.
- ↑ "The Nobel Prize in Physiology or Medicine 1980 - Speed Read". https://www.nobelprize.org/nobel_prizes/medicine/laureates/1980/speedread.html.
- ↑ Kasahara, M. (2000). Major Histocompatibility Complex: Evolution, Structure, and Function. New York: Springer. ISBN 978-4-431-70276-4.
- ↑ Delves, Peter J. (January 2017). "Human Leukocyte Antigen (HLA) System". http://www.merckmanuals.com/professional/immunology-allergic-disorders/biology-of-the-immune-system/human-leukocyte-antigen-hla-system.
- ↑ "T-lymphocyte recognition of antigen in association with gene products of the major histocompatibility complex". Annual Review of Immunology 3 (1): 237–61. 1985-01-01. doi:10.1146/annurev.iy.03.040185.001321. PMID 2415139.
- ↑ "The major histocompatibility complex in transplantation". Journal of Transplantation 2012: 842141. 2012. doi:10.1155/2012/842141. PMID 22778908.
- ↑ "The Major Histocompatibility Complex and Its Functions". Immunobiology: The Immune System in Health and Disease (5th ed.). Elsevier España. 2001. ISBN 978-0-8153-3642-6. https://www.ncbi.nlm.nih.gov/books/NBK27156.
- ↑ "Major histocompatibility complex genomics and human disease". Annual Review of Genomics and Human Genetics 14: 301–23. 2013. doi:10.1146/annurev-genom-091212-153455. PMID 23875801.
- ↑ 17.0 17.1 "HLA Mismatching Strategies for Solid Organ Transplantation - A Balancing Act". Frontiers in Immunology 7: 575. 2016. doi:10.3389/fimmu.2016.00575. PMID 28003816.
- ↑ Cruz-Tapias, Paola; Castiblanco, John; Anaya, Juan-Manuel (2013-07-18). Major histocompatibility complex: Antigen processing and presentation. El Rosario University Press. https://www.ncbi.nlm.nih.gov/books/NBK459467/.
- ↑ "HLA matching for kidney transplantation". Human Immunology 65 (12): 1489–505. December 2004. doi:10.1016/j.humimm.2004.06.008. PMID 15603878.
- ↑ "Haploidentical Hematopoietic Stem Cell Transplantation: A Global Overview Comparing Asia, the European Union, and the United States". Biology of Blood and Marrow Transplantation 22 (1): 23–6. January 2016. doi:10.1016/j.bbmt.2015.11.001. PMID 26551633.
- ↑ Immune response and the eye: in memoriam J. Wayne Streilein. Niederkorn, J. Y. (Jerry Y.), 1946-, Kaplan, Henry J., Streilein, J. Wayne. (2nd, rev. ed.). Basel: Karger. 2007. ISBN 9783805581875. OCLC 85243138.
- ↑ "Transplanting the elderly: Balancing aging with histocompatibility". Transplantation Reviews 29 (4): 205–11. October 2015. doi:10.1016/j.trre.2015.08.003. PMID 26411382.
- ↑ "Immunosuppressive drugs after solid organ transplantation". The Netherlands Journal of Medicine 71 (6): 281–9. July 2013. PMID 23956308.
- ↑ "Role of major histocompatibility complex variation in graft-versus-host disease after hematopoietic cell transplantation". F1000Research 6: 617. 2017-05-03. doi:10.12688/f1000research.10990.1. PMID 28529723.
- ↑ 25.0 25.1 25.2 Chandraker, Anil; Sayegh, Mohamed H. (2012). Core concepts in renal transplantation. New York: Springer Science+Business Media, LLC. pp. 1960. ISBN 9781461400073. OCLC 768245800.
External links
- Histocompatibility at the US National Library of Medicine Medical Subject Headings (MeSH)
 |
