Biology:Mercury transporter
| Transporter MerF | |||||||||
|---|---|---|---|---|---|---|---|---|---|
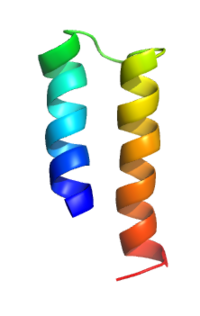 | |||||||||
| Identifiers | |||||||||
| Symbol | MerF | ||||||||
| Pfam | PF11431 | ||||||||
| InterPro | IPR021091 | ||||||||
| TCDB | 1.A.72 | ||||||||
| OPM superfamily | 218 | ||||||||
| OPM protein | 2lj2 | ||||||||
| |||||||||
The mercury transporter superfamily (TC# 1.A.72) is a family of transmembrane bacterial transporters of mercury ions. The common origin of all Mer superfamily members has been established.[2] The common elements between family members are included in TMSs 1-2. A representative list of the subfamilies and proteins that belong to those subfamilies is available in the Transporter Classification Database.
Subfamilies
- 1.A.72.1: The MerF Mercuric Ion (Hg²⁺) Uptake (MerF) Family
- 1.A.72.2: The MerH Mercuric Ion (Hg²⁺) Permease (MerH) Family
- 1.A.72.3: The MerTP Mercuric Ion (Hg²⁺) Permease (MerTP) Family
- 1.A.72.4: The MerC Mercuric Ion (Hg²⁺) Permease (MerC) Family
- 1.A.72.5: The MerE Mercuric Ion (Hg²⁺) Permease (MerE) Family
Transport Reaction
The transport reaction catalyzed by Mer Superfamily members is:
Hg2+ or methyl-Hg2+ (out) → Hg2+ or methyl-Hg2+ (in)
MerF
The MerF protein encoded on plasmid pMER327/419 is an 81 residue polypeptide with two putative TMSs.[3] It catalyzes uptake of Hg2+ in preparation for reduction by mercuric reductase. The MerF gene is found on mercury resistant plasmids from many gram-negative bacteria, but the sequence of the protein from these plasmids is the same. Limited sequence similarity is shown with the first two TMSs of MerT (TC# 1.A.72.3) and MerC (TC# 1.A.72.4). MerF has two vicinal pairs of cysteine residues which are involved in the transport of Hg(II) across the membrane and are exposed to the cytoplasm.[4] Some members of the MerF family have been designated MerH.[5]
Crystal structures
PDB: 1WAZ, 2H3O, 2LJ2, 2M67, 2MOZ
MerTP
The MerTP permeases catalyze uptake into bacterial cells of Hg2+ in preparation for its reduction by the MerA mercuric reductase. The Hgo produced by MerA is volatile and passively diffuses out of the cell. The merT and merP genes are found on mercury resistance plasmids and transposons of gram-negative and gram-positive bacteria but are also chromosomally encoded in some bacteria. MerT consists of about 130 amino acids and has 3 transmembrane helical segments.[6] Operon analyses have been reported.[3][7][8]
MerP
MerP is a periplasmic Hg2+-binding receptor of about 70-80 amino acyl residues, synthesized with a cleavable N-terminal leader. It is homologous to the N-terminal heavy metal binding domains of the copper-and cadmium-transporting P-type ATPases. The 3-D structure of MerP from Ralstonia metallidurans has been solved to 2 Å resolution (PDB: 1OSD).[9][10] It is 91 amino acyl residues (aas) long with its leader sequence, is monomeric, and binds a single Hg2+ ion. Hg2+ is bound to a sequence GMTCXXC found in metallochaperones as well as metal-transporting ATPases. The fold is βαββαβ, called the ''ferridoxin-like fold''.
MerT
MerT homologues have been identified in which the 3 TMS MerT is fused to a MerP ''heavy metal associated'' (HMA) domain, possibly via a linker region that includes a fourth TMS (see 1.A.72.3.3). HMA domains of ~30 aas are found in MerP, copper chaperone proteins, mercuric reductase, and at the N-termini of both copper and heavy metal P-type ATPases, sometimes in multiple copies.[11]
MerC
The MerC protein encoded on the IncJ plasmid pMERPH of the Shewanella putrefaciens mercuric resistance operon is 137 amino acids in length and possesses four putative transmembrane α-helical spanners (TMSs). It has been shown to bind and take up Hg2+ ions. merC genes are encoded on several plasmids of gram-negative bacteria and may also be chromosomally encoded. MerC proteins are homologous to other bacterial Hg2+ bacterial transporters.[2][12][13][14]
MerE
See Kiyono, Masako; Sone, Yuka; Nakamura, Ryosuke; Pan-Hou, Hidemitsu; Sakabe, Kou (2009-04-02). "The MerE protein encoded by transposon Tn21 is a broad mercury transporter in Escherichia coli". FEBS Letters 583 (7): 1127–1131. doi:10.1016/j.febslet.2009.02.039. ISSN 1873-3468. PMID 19265693.
References
- ↑ De Angelis, A. A.; Howell, S. C.; Nevzorov, A. A.; Opella, S. J. (2006). "Structure Determination of a Membrane Protein with Two Trans-membrane Helices in Aligned Phospholipid Bicelles by Solid-State NMR Spectroscopy". Journal of the American Chemical Society 128 (37): 12256–12267. doi:10.1021/ja063640w. PMID 16967977.
- ↑ 2.0 2.1 Mok, Timothy; Chen, Jonathan S.; Shlykov, Maksim A.; Jr, Milton H. Saier (2012-06-02). "Bioinformatic Analyses of Bacterial Mercury Ion (Hg2+) Transporters" (in en). Water, Air, & Soil Pollution 223 (7): 4443–4457. doi:10.1007/s11270-012-1208-3. ISSN 0049-6979. Bibcode: 2012WASP..223.4443M.
- ↑ 3.0 3.1 Barkay, Tamar; Miller, Susan M.; Summers, Anne O. (2003-06-01). "Bacterial mercury resistance from atoms to ecosystems". FEMS Microbiology Reviews 27 (2–3): 355–384. doi:10.1016/s0168-6445(03)00046-9. ISSN 0168-6445. PMID 12829275.
- ↑ "NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system". Biochemistry 44 (13): 5196–206. April 2005. doi:10.1021/bi048095v. PMID 15794657.
- ↑ Wilson, J. R.; Leang, C.; Morby, A. P.; Hobman, J. L.; Brown, N. L. (2000-04-21). "MerF is a mercury transport protein: different structures but a common mechanism for mercuric ion transporters?". FEBS Letters 472 (1): 78–82. doi:10.1016/s0014-5793(00)01430-7. ISSN 0014-5793. PMID 10781809.
- ↑ Schué, Mathieu; Dover, Lynn G.; Besra, Gurdyal S.; Parkhill, Julian; Brown, Nigel L. (2009-01-01). "Sequence and analysis of a plasmid-encoded mercury resistance operon from Mycobacterium marinum identifies MerH, a new mercuric ion transporter". Journal of Bacteriology 191 (1): 439–444. doi:10.1128/JB.01063-08. ISSN 1098-5530. PMID 18931130.
- ↑ Miller, S. M. (1999-01-01). "Bacterial detoxification of Hg(II) and organomercurials". Essays in Biochemistry 34: 17–30. doi:10.1042/bse0340017. ISSN 0071-1365. PMID 10730186.
- ↑ Velasco, A.; Acebo, P.; Flores, N.; Perera, J. (1999-01-01). "The mer operon of the acidophilic bacterium Thiobacillus T3.2 diverges from its Thiobacillus ferrooxidans counterpart". Extremophiles: Life Under Extreme Conditions 3 (1): 35–43. doi:10.1007/s007920050097. ISSN 1431-0651. PMID 10086843.
- ↑ Serre, Laurence; Rossy, Emmanuel; Pebay-Peyroula, Eva; Cohen-Addad, Claudine; Covès, Jacques (2004-05-21). "Crystal structure of the oxidized form of the periplasmic mercury-binding protein MerP from Ralstonia metallidurans CH34". Journal of Molecular Biology 339 (1): 161–171. doi:10.1016/j.jmb.2004.03.022. ISSN 0022-2836. PMID 15123428.
- ↑ Qian, H.; Sahlman, L.; Eriksson, P. O.; Hambraeus, C.; Edlund, U.; Sethson, I. (1998-06-30). "NMR solution structure of the oxidized form of MerP, a mercuric ion binding protein involved in bacterial mercuric ion resistance". Biochemistry 37 (26): 9316–9322. doi:10.1021/bi9803628. ISSN 0006-2960. PMID 9649312.
- ↑ Morby, A. P.; Hobman, J. L.; Brown, N. L. (1995-07-01). "The role of cysteine residues in the transport of mercuric ions by the Tn501 MerT and MerP mercury-resistance proteins". Molecular Microbiology 17 (1): 25–35. doi:10.1111/j.1365-2958.1995.mmi_17010025.x. ISSN 0950-382X. PMID 7476206.
- ↑ Chugh, Pauline; Bradel-Tretheway, Birgit; Monteiro-Filho, Carlos MR; Planelles, Vicente; Maggirwar, Sanjay B; Dewhurst, Stephen; Kim, Baek (2008-01-31). "Akt inhibitors as an HIV-1 infected macrophage-specific anti-viral therapy". Retrovirology 5: 11. doi:10.1186/1742-4690-5-11. ISSN 1742-4690. PMID 18237430.
- ↑ Harkema, J. R.; Hotchkiss, J. A. (1992-08-01). "In vivo effects of endotoxin on intraepithelial mucosubstances in rat pulmonary airways. Quantitative histochemistry.". The American Journal of Pathology 141 (2): 307–317. ISSN 0002-9440. PMID 1497089.
- ↑ Yamaguchi, Ai; Tamang, Dorjee G.; Jr, Milton H. Saier (2007-02-06). "Mercury Transport in Bacteria" (in en). Water, Air, and Soil Pollution 182 (1–4): 219–234. doi:10.1007/s11270-007-9334-z. ISSN 0049-6979. Bibcode: 2007WASP..182..219Y.
 |

