Biology:NBEAL1
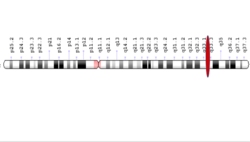
NBEAL1 is a protein that in humans is encoded by the NBEAL1 gene.[1] It is found on chromosome 2q33.2 of Homo sapiens.
| Neurobeachin-like protein 1 | |
|---|---|
| Identifiers | |
| Symbol | NBEAL1 |
| Alt. names | ALS2CR16 ALS2CR17 |
| NCBI gene | 65065 |
| HGNC | 20681 |
| OMIM | 609816 |
| UniProt | Q6ZS30 |
| Other data | |
| Locus | Chr. 2 q33.2 |
Through the different domains of this protein, the function of NBEAL1 is predicted to be involved in the following cellular mechanisms: vesicle trafficking, membrane dynamics, receptor signaling, pre-mRNA processing, signal transduction and cytoskeleton assembly.[2][3][4] NBEAL1 is also known as Amytorophic Lateral Sclerosis 2 Chromosomal Region, ALS2CR16 and ALS2CR17.[1]
Protein Properties
Transcript
The mRNA for this protein consists of 9058 base pairs in a linear sequence with the coding sequence begins at base pair number 334 and extends until base pair number 8418.[5] The translated protein is a total 56 exons that constitute a final length of 2694 amino acids.[6] There are currently 9 known isoforms within humans.[3]
Domains
Neurobeachin-like1 contains five domains: DUF4704, DUF4800, PH_BEACH, Beach, and WD40 repeats.[6]
DUF4704
DUF4704 is a domain of unknown function. While the function of this domain is unknown, it is conserved within neurobeachin proteins in eukaryotes.[4] It begins at amino acid 859 and spans until number 1115.[3]
DUF4800
DUF4800 is a domain of unknown function. It begins at amino acid 1580, spanning until 1833.[3] While it is uncharacterized in function, it is found within eukaryotes.[7]
PH_BEACH
Spanning from amino acid 1886 until amino acid 1983, this domain is referred to as a Pleckstrin Homology domain in the BEACH domain.[8] It has a PH because the fold of this domain is similar to the PH domain, but is not identical in the sequence of the canonical PH domains. The PH_BEACH domain is not able to bind phospholipids.[9]
Beach
The Beige and Chediak-Higashi (BEACH) domain is one of the most significant domains within this protein. This domain is highly conserved roughly 280 amino acid domain, present in nine different human BEACH domains.[10] It located after the PH_BEACH domain in the sequence. While not much is understood on the exact function of BDCP proteins within the BEACH domain, it is known that they serve many purposes within cellular mechanisms: vesicular transport, apoptosis, membrane dynamics and receptor signaling.[10] This protein family is of great clinical importance currently because mutations in this domain have been identified in multiple human disorders. For example, neurobeachin-like1 is upregulated in glioma: as the pathological grade of the glioma increases, the expression of neurobeachin-like1 is decreased.[2] In NBEAL1, this follows the PH_BEACH domain, beginning at amino acid 2005 and ending at amino acid 2284.[3]
WD40
NBEAL1 has one WD40 domain within NBEAL1. From amino acid 2409 to 2682 is the entire WD40 domain. Within the domain, from 2406 to 2439, there is a structural motif WD40 repeat. The WD40 domain is found in a number of eukaryotic proteins that have multiple functions. These include, but are not limited to, adaptor/regulatory modules in signal transduction, pre-mRNA processing, and cytoskeleton assembly.[3]
Properties
- Molecular weight of 307.3 kilodaltons[11]
- Predicted to be localized to the mitochondria[12]
Structure
Secondary
The secondary structure of NBEAL1 is predicted to be a combination of alpha helices, beta sheets and random coils.[13]
Tertiary

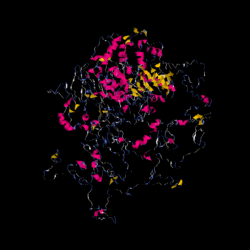
I-TASSER was used to predict a 3D structure of NBEAL1.[14] Since NBEAL1 is longer in amino acid length than allowed for input, it was split in half to predict the structure of the whole protein.
Post-Translational Modifications
The following document illustrates the different post-translational modifications.
Expression
Using the EST abundance profile through Unigene, NBEAL1 expression was discovered based on both body sites and health states.[15] NBEAL1 shows expression in the brain, embryonic tissue, eye, intestine, kidney, liver, lung, mammary glands, ovaries, pancreas, pharynx, placenta, prostate, skin, stomach, testis, thyroid, and trachea. Based on transcripts per million, expression is highest in the stomach at 62 transcripts per million, with pancreas and trachea being next with their transcripts per million being 37 and 38, respectively. The lowest transcripts per million in the brain, eye, placenta and testis, all at 4 per million. When looking at the breakdown by different health states, NBEAL1 is highly expressed in multiple tumors.[15] Again, the abundance was highest in gastrointestinal tumors, correlating to the high expression of NBEAL1 within the stomach. However, NBEAL1 expression is not seen in pancreatic tumors, which may signify something about its function within the pancreas. The abundance also differs in developmental stages, the highest being the fetal stage with 21 transcripts per million and the adult at 14 transcripts per million.
Function
The function of NBEAL1 is not yet well understood by the scientific community. However, given the function of the different domains and disease associations, it is predicted that the NBEAL1 protein may be involved in a variety of functions. As of now they include, but are not limited to, protein-protein interactions, vesicle trafficking, membrane dynamics, receptor signaling, apoptosis, adaptor/regulatory modules in signal transduction, pre-mRNA processing, and cytoskeleton assembly.[3][2]
Clinical Significance
This protein has been associated with NBEAL1 are Amyotrophic Lateral Sclerosis, Juvenile and Adenocarcinoma,[1] although the function in these diseases has not yet been identified.
Homology
Neurobeachin-like1 is a highly conserved protein. It has orthologs found in many life forms, including but not limited to: reptiles, birds, amphibians, mammals, fish, and a few invertebrates. The following table presents some of the orthologs found using searches in BLAST[16] and BLAT.[17]
| Scientific Name | Common Name | Accession Number | Sequence Length | Percent Identity |
|---|---|---|---|---|
| Homo sapiens | Human | NP_001107604.1 | 2694 | - |
| Pan troglodytes | Chimpanzee | XP_525997.3 | 2694 | 99 |
| Gorilla gorilla gorilla | Western Lowland Gorilla | XP_018878299.1 | 2694 | 99 |
| Mus musculus | Mouse | NP_77560 | 2688 | 98 |
| Cerocebus tays | Sooty angabey | XP_011903312 | 2678 | 97 |
| Canis lupus familiaris | Dog | XP_545603.3 | 2693 | 93 |
| Ailuropoda melanoleuca | Giant Panda | XP_019655126.1 | 2693 | 93 |
| Trichechus manatus latirostris | West Indian Manatee | XP_004378299 | 2682 | 93 |
| Tursiops truncatus | Common Bottlenose Dolphin | XP_019794654.1 | 2682 | 92.7 |
| Eptesicus cuscus | Big Brown Bat | XP_008144758.1 | 2722 | 92 |
| Zonotrichia albicollis | White Throated Sparrow | XP_014120514.1 | 2707 | 80 |
| Gallus gallus | Chicken | XP_004942730.1 | 2725 | 78.7 |
| Python bivattatus | Burmese Python | XP_007422078.1 | 2687 | 79 |
| Xenopus tropicalis | Western Clawed Frog | XP_012826463.1 | 2687 | 74 |
| Callorhinchus milii | Australian Ghostshark | XP_007888887.1 | 2749 | 70.2 |
| Danio rerio | Zebrafish | XP_009300392 | 2723 | 66.3 |
| Octopus bimaculoides | California two-spot octopus | XP_014777916.1 | 2584 | 41.2 |
| Daphnia magna | Planktonic crustacean | KZS03729 | 2734 |
34.4 |
| Drosophila busckii | Fruit fly | XP_017842328.1 | 2722 | 34 |

Paralogs
According to GeneCards, NBEAL1 has a few paralogs: NBEAL2, WDFY3, NBEA, LRBA, Lysosomal trafficking regulator (LYST), and WDFY3.[19] The table below summarizes the paralogs of NBEAL1.
| Gene Name | Species | Accession Number | Sequence Length | Percent Identity |
|---|---|---|---|---|
| NBEAL2 | Homo sapiens | NP_055990 | 2754 | 46 |
| NBEA | Homo sapiens | NP_056483.3 | 2946 | 22.8 |
| LRBA | Homo sapiens | NP_996717.2 | 2863 | 22.5 |
| WDFY3 | Homo sapiens | XP_016863397.1 | 3544 | 21.8 |
| LYST | Homo sapiens | NP_001288294.1 | 3801 | 19.3 |
References
- ↑ 1.0 1.1 1.2 Database, GeneCards Human Gene. "NBEAL1 Gene - GeneCards | NBEL1 Protein | NBEL1 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=NBEAL1#orthologs.
- ↑ 2.0 2.1 2.2 "Identification and characterization of NBEAL1, a novel human neurobeachin-like 1 protein gene from fetal brain, which is up regulated in glioma". Brain Research. Molecular Brain Research 125 (1–2): 147–55. June 2004. doi:10.1016/j.molbrainres.2004.02.022. PMID 15193433.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 "homo sapiens NBEAL1 - Protein - NCBI". https://www.ncbi.nlm.nih.gov/protein/?term=homo+sapiens+NBEAL1.
- ↑ 4.0 4.1 "SEL-2, the C. elegans neurobeachin/LRBA homolog, is a negative regulator of lin-12/Notch activity and affects endosomal traffic in polarized epithelial cells". Development 134 (4): 691–702. February 2007. doi:10.1242/dev.02767. PMID 17215302.
- ↑ "Homo sapiens neurobeachin like 1 (NBEAL1), mRNA - Nucleotide - NCBI". https://www.ncbi.nlm.nih.gov/nuccore/224451123.
- ↑ 6.0 6.1 "neurobeachin-like protein 1 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/224451124.
- ↑ "Pfam: Family: DUF4800 (PF16057)". http://pfam.xfam.org/family/DUF4800.
- ↑ "NCBI CDD Conserved Protein Domain PH_BEACH" (in en). https://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=275391.
- ↑ "Crystal structure of the PH-BEACH domains of human LRBA/BGL". Biochemistry 43 (47): 14873–80. November 2004. doi:10.1021/bi049498y. PMID 15554694.
- ↑ 10.0 10.1 "The BEACH is hot: a LYST of emerging roles for BEACH-domain containing proteins in human disease". Traffic 14 (7): 749–66. July 2013. doi:10.1111/tra.12069. PMID 23521701.
- ↑ Kramer, Jack (1990). "Molecular Weight". http://seqtool.sdsc.edu/CGI/BW.cgi#!.[yes|permanent dead link|dead link}}]
- ↑ Nakai and Horton (1997). "PSORTII". http://www.psort.org.
- ↑ "ExPASy: SIB Bioinformatics Resource Portal - Home" (in en-US). http://www.expasy.org.
- ↑ "I-TASSER server for protein structure and function prediction". http://zhanglab.ccmb.med.umich.edu/I-TASSER/.
- ↑ 15.0 15.1 Group, Schuler. "EST Profile - Hs.648846". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.648846.
- ↑ "BLAST: Basic Local Alignment Search Tool". https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "Genome Browser FAQ". https://genome.ucsc.edu/FAQ/FAQblat.html.
- ↑ Workbench, NCSA Biology. "SDSC Biology Workbench". http://seqtool.sdsc.edu/.
- ↑ Database, GeneCards Human Gene. "NBEAL1 Gene - GeneCards | NBEL1 Protein | NBEL1 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=NBEAL1#paralogs.
Further reading
- "A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2". Nature Genetics 29 (2): 166–73. October 2001. doi:10.1038/ng1001-166. PMID 11586298.
- "Identification and characterization of NBEAL1, a novel human neurobeachin-like 1 protein gene from fetal brain, which is up regulated in glioma". Brain Research. Molecular Brain Research 125 (1–2): 147–55. June 2004. doi:10.1016/j.molbrainres.2004.02.022. PMID 15193433.
 |
