Biology:Paenibacillus vortex
| Paenibacillus vortex | |
|---|---|
| File:Paenibacillus fig 1.tif | |
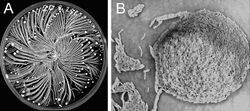
| Figure 1: Colony organization of the P. vortex bacteria when grown on 15g/L peptone and 2.25% (w/v) agar for four days. The bright yellow dots are the vortices. The colonies were grown in a Petri dish size 8.8cm and stained with Coomassie dyes (Brilliant Blue). The colors were inverted to emphasize higher densities using the brighter shades of yellow. | |
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Binomial name | |
| Paenibacillus vortex | |
| Synonyms | |
|
Bacillus vortex
Ash et al. 1994 | |
Paenibacillus vortex is a species of pattern-forming bacteria, first discovered in the early 1990s by Eshel Ben-Jacob's group at Tel Aviv University.[1] It is a social microorganism that forms colonies with complex and dynamic architectures. P. vortex is mainly found in heterogeneous and complex environments, such as the rhizosphere, the soil region directly influenced by plant roots.
The genus Paenibacillus comprises facultative anaerobic, endospore-forming bacteria originally included within the genus Bacillus and then reclassified as a separate genus in 1993.[2] Bacteria in the genus have been detected in a variety of environments such as: soil, water, vegetable matter, forage and insect larvae, as well as clinical samples.[3][4][5][6] Paenibacillus spp., including P. vortex, produce extracellular enzymes that catalyze a variety of synthetic reactions in industrial, agricultural and medical applications.[7][8][9] Various Paenibacillus spp. also produce antimicrobial substances that can affect micro-organisms such as fungi, in addition to soil and plant pathogenic bacteria.[10][11][12]
Social Motility
Paenibacillus vortex possesses advanced social motility employing cell-cell attractive and repulsive chemotactic signalling and physical links. When grown on soft surfaces (e.g. agar), the collective motility is reflected by the formation of foraging swarms[13] that act as arms sent out in search of food. These swarms have an aversion to crossing each other’s trail and collectively change direction when food is sensed. The “swarming intelligence” P. vortex, is further marked by the fact that of the swarms can even split and reunite when detecting scattered patches of nutrients.[13]
Pattern Formation and Social Behaviors
thumb|alt= alt text|Figure 2: Scanning electron microscope (SEM) observation of P. vortex illustrating a typical bacteria arrangement in the center of a vortex. Notable, that each individual bacterium is curved. Scale bar in is 5µm. P. vortex is a social microorganism: when grown on under growth conditions that mimic natural environments such as hard surfaces it forms colonies of 109-1012 cells with remarkably complex and dynamic architectures (Figure 1).[1][14][15] Being part of a large cooperative, the bacteria can better compete for food resources and be protected against antibacterial assaults.[13][14] Under laboratory growth conditions, similar to other social bacteria, P. vortex colonies behave much like a multi-cellular organism, with cell differentiation and task distribution.[16][17][18][19] P. vortex is marked by its ability to generate special aggregates of dense bacteria that are pushed forward by repulsive chemotactic signals sent from the cells at the back.[15][20][21][22][23][24] These rotating aggregates termed vortices (Figure 2), pave the way for the colony to expand. The vortices serve as building blocks of colonies with special modular organization (Figure 1). Accomplishing such intricate cooperative ventures requires sophisticated cell-cell communication,[14][19][25][26][27] including semantic and pragmatic aspects of linguistics.[19] Communicating with each other using a variety of chemical signals, bacteria exchange information regarding population size, a myriad of individual environmental measurements at different locations, their internal states and their phenotypic and epigenetic adjustments. The bacteria collectively sense the environment and execute distributed information processing to glean and assess relevant information.[14][19][28] The information is then used by the bacteria for reshaping the colony while redistributing tasks and cell epigenetic differentiations, for collective decision-making and for turning on and off defense and offense mechanisms needed to thrive in competitive environments, faculties that can be perceived as social intelligence of bacteria.[19]
Genome Sequence of the Paenibacillus vortex
The genome sequence of the P. vortex [29] is now available [GenBank: ADHJ00000000). The genome was sequenced by a hybrid approach using 454 Life Sciences and Illumina, achieving a total of 289X coverage, with 99.8% sequence identity between the two methods. The sequencing results were validated using a custom designed Agilent microarray expression chip submitted to EMBL-EBI [ArrayExpress: E-MEXP-3019 which represented the coding and the non-coding regions. Analysis of the P. vortex genome revealed 6,437 open reading frames (ORFs) and 73 non-coding RNA genes. The analysis also unveiled the P. vortex potential to produce a wealth of enzymes and proteases as well as a great variety of antimicrobial substances that affect a wide range of microorganisms. The possession of these advanced defense and offense strategies render Paenibacillus vortex as a rich source of useful genes for agricultural, medical, industrial and biofuel applications.
Comparative Genomics and Social-IQ Score
Comparative genomic analysis revealed that bacteria successful in heterogeneous and competitive environments often contain extensive signal transduction and regulatory networks.[30][31][32] Detailed comparative genomic analysis with a dataset of 500 complete bacterial genomes revealed that P. vortex has the third highest number of signal transduction genes, slightly below two other Paenibacillus species, Paenibacillus sp. JDR-2 and Paenibacillus sp. Y412MC10.[29] The comparative genomic analysis further revealed that these three Paenibacillus species also have the highest “Social-IQ” score among all 500 sequenced bacteria, over 3 standard deviations higher than average. The score is based on the number of genes which afford bacteria abilities to communicate and process environmental information (two-component and transcription-factor genes), to make decisions and to synthesize offensive (toxic) and defensive (neutralizing) agents as needed during chemical warfare with other microorganisms.[29] Defined this way, the Social-IQ score provides a measure of the genome capacity for social intelligence, hence it helps realizing social intelligence of bacteria.
References
- ↑ 1.0 1.1 "Evolution of complexity during growth of bacterial colonies". NATO Advanced Research Workshop (Santa Fe, USA: Addison-Wesley Publishing Company): 619–633. 1995.
- ↑ Ash C, Priest FG, Collins MD: Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie van Leeuwenhoek 1993, 64:253-260.
- ↑ Lal, Sadhana; Tabacchioni, Silvia (2009). "Ecology and biotechnological potential of Paenibacillus polymyxa: a minireview". Indian Journal of Microbiology (Springer Science and Business Media LLC) 49 (1): 2–10. doi:10.1007/s12088-009-0008-y. ISSN 0046-8991. PMID 23100748.
- ↑ McSpadden Gardener, Brian B. (2004). "Ecology of Bacillus and Paenibacillus spp. in Agricultural Systems". Phytopathology (Scientific Societies) 94 (11): 1252–1258. doi:10.1094/phyto.2004.94.11.1252. ISSN 0031-949X. PMID 18944463.
- ↑ Montes, M. J. (2004-09-01). "Paenibacillus antarcticus sp. nov., a novel psychrotolerant organism from the Antarctic environment". International Journal of Systematic and Evolutionary Microbiology (Microbiology Society) 54 (5): 1521–1526. doi:10.1099/ijs.0.63078-0. ISSN 1466-5026. PMID 15388704.
- ↑ Ouyang J, Pei Z, Lutwick L, Dalal S, Yang L, Cassai N, Sandhu K, Hanna B, Wieczorek RL, Bluth M, Pincus MR: Case report: Paenibacillus thiaminolyticus: a new cause of human infection, inducing bacteremia in a patient on hemodialysis. Ann Clin Lab Sci 2008, 38:393-400.
- ↑ Konishi, J.; Maruhashi, K. (2003-09-01). "2-(2'-Hydroxyphenyl)benzene sulfinate desulfinase from the thermophilic desulfurizing bacterium Paenibacillus sp. strain A11-2: purification and characterization". Applied Microbiology and Biotechnology (Springer Science and Business Media LLC) 62 (4): 356–361. doi:10.1007/s00253-003-1331-6. ISSN 0175-7598. PMID 12743754.
- ↑ Raza W, Yang W, Shen QR: Paenibacillus polymyxa: Antibiotics, Hydrolytic Enzymes and Hazard Assessment. J Plant Pathol 2008, 90:419-430.
- ↑ Watanapokasin RY, Boonyakamol A, Sukseree S, Krajarng A, Sophonnithiprasert T, Kanso S, Imai T: Hydrogen production and anaerobic decolorization of wastewater containing Reactive Blue 4 by a bacterial consortium of Salmonella subterranea and Paenibacillus polymyxa. Biodegradation 2009, 20:411-418.
- ↑ Dijksterhuis J, Sanders M, Gorris LG, Smid EJ: Antibiosis plays a role in the context of direct interaction during antagonism of Paenibacillus polymyxa towards Fusarium oxysporum. J Appl Microbiol 1999, 86:13-21.
- ↑ Girardin H, Albagnac C, Dargaignaratz C, Nguyen-The C, Carlin F: Antimicrobial activity of foodborne Paenibacillus and Bacillus spp. against Clostridium botulinum. J Food Prot 2002, 65:806-813.
- ↑ von der Weid I, Alviano DS, Santos AL, Soares RM, Alviano CS, Seldin L: Antimicrobial activity of Paenibacillus peoriae strain NRRL BD-62 against a broad spectrum of phytopathogenic bacteria and fungi. J Appl Microbiol 2003, 95:1143-1151.
- ↑ 13.0 13.1 13.2 Ingham CJ, Ben-Jacob E: Swarming and complex pattern formation in Paenibacillus vortex studied by imaging and tracking cells. BMC Microbiol 2008, 8:36.
- ↑ 14.0 14.1 14.2 14.3 Ben-Jacob E: Bacterial self-organization: co-enhancement of complexification and adaptability in a dynamic environment. Phil Trans R Soc Lond A 2003, 361:1283-1312.
- ↑ 15.0 15.1 Ben-Jacob E, Cohen I, Gutnick DL: Cooperative organization of bacterial colonies: from genotype to morphotype. Annu Rev Microbiol 1998, 52:779-806.
- ↑ Aguilar C, Vlamakis H, Losick R, Kolter R: Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol 2007, 10:638-643.
- ↑ Dunny GM, Brickman TJ, Dworkin M: Multicellular behavior in bacteria: communication, cooperation, competition and cheating. Bioessays 2008, 30:296-298.
- ↑ Shapiro JA, Dworkin M: Bacteria as multicellular organisms. 1st edn: Oxford University Press, USA; 1997.
- ↑ 19.0 19.1 19.2 19.3 19.4 Ben-Jacob E, Becker I, Shapira Y, Levine H: Bacterial linguistic communication and social intelligence. Trends Microbiol 2004, 12:366-372.
- ↑ Ben-Jacob E: From snowflake formation to growth of bacterial colonies II: Cooperative formation of complex colonial patterns. Contem Phys 1997, 38:205 - 241.
- ↑ Ben-Jacob E, Cohen I: Cooperative formation of bacterial patterns. In Bacteria as Multicellular Organisms Edited by Shapiro JA, Dworkin M. New York: Oxford University Press; 1997: 394-416
- ↑ Ben-Jacob E, Cohen I, Czirók A, Vicsek T, Gutnick DL: Chemomodulation of cellular movement, collective formation of vortices by swarming bacteria, and colonial development. Physica A 1997, 238:181-197.
- ↑ Cohen I, Czirok A, Ben-Jacob E: Chemotactic-based adaptive self-organization during colonial development. Physica A 1996, 233:678-698.
- ↑ Czirók, András; Ben-Jacob, Eshel; Cohen, Inon; Vicsek, Tamás (1996-08-01). "Formation of complex bacterial colonies via self-generated vortices". Physical Review E (American Physical Society (APS)) 54 (2): 1791–1801. doi:10.1103/physreve.54.1791. ISSN 1063-651X. PMID 9965259. Bibcode: 1996PhRvE..54.1791C.
- ↑ Bassler BL, Losick R: Bacterially speaking. Cell 2006, 125:237-246.
- ↑ Bischofs IB, Hug JA, Liu AW, Wolf DM, Arkin AP: Complexity in bacterial cell-cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proc Natl Acad Sci U S A 2009, 106:6459-6464.
- ↑ Kolter R, Greenberg EP: Microbial sciences: the superficial life of microbes. Nature 2006, 441:300-302.
- ↑ Dwyer DJ, Kohanski MA, Collins JJ: Networking opportunities for bacteria. Cell 2008, 135:1153-1156.
- ↑ 29.0 29.1 29.2 Sirota-Madi A, Olender T, Helman Y, Ingham C, Brainis I, Roth D, Hagi E, Brodsky L, Leshkowitz D, Galatenko V, et al: Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments. BMC Genomics, 11:710.
- ↑ Alon U: An Introduction to Systems Biology: Design Principles of Biological circuits. London, UK: CRC Press; 2006.
- ↑ Galperin MY, Gomelsky M: Bacterial Signal Transduction Modules: from Genomics to Biology. ASM News 2005, 71:326-333.
- ↑ Whitworth DE, Cock PJ: Two-component systems of the myxobacteria: structure, diversity and evolutionary relationships. Microbiology 2008, 154:360-372.
External links
- Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments.
- Prof. Eshel Ben-Jacob's Home Page
- Realizing Social Intelligence of Bacteria
- Bacterial Art
Wikidata ☰ Q7124085 entry
 |