Biology:Phase variation
In biology, phase variation[1] is a method for dealing with rapidly varying environments without requiring random mutation. It involves the variation of protein expression, frequently in an on-off fashion, within different parts of a bacterial population. As such the phenotype can switch at frequencies that are much higher (sometimes >1%) than classical mutation rates. Phase variation contributes to virulence by generating heterogeneity. Although it has been most commonly studied in the context of immune evasion, it is observed in many other areas as well and is employed by various types of bacteria, including Salmonella species.
Salmonella use this technique to switch between different types of the protein flagellin. As a result, flagella with different structures are assembled. Once an adaptive response has been mounted against one type of flagellin, or if a previous encounter has left the adaptive immune system ready to deal with one type of flagellin, switching types renders previously high-affinity antibodies, T-cell receptors, and B-cell receptors ineffective against the flagella.
Site-specific recombination
Site-specific recombinations are usually short and occur at a single target site within the recombining sequence. For this to occur there are typically one or more cofactors (to name a few: DNA-binding proteins and the presence or absence of DNA binding sites) and a site-specific recombinase.[2] There is a change in orientation of the DNA that will affect gene expression or the structure of the gene product.[3] This is done by changing the spatial arrangement of the promoter or the regulatory elements.[2]
Inversion
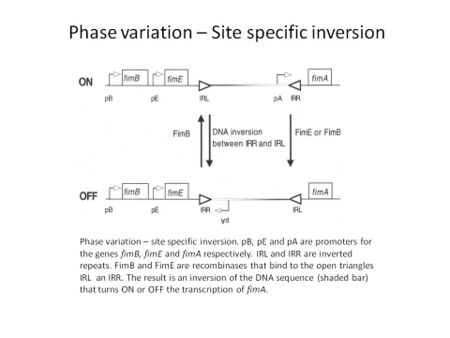
Through the utilization of specific recombinases, a particular DNA sequence is inverted, resulting in an ON to OFF switch and vice versa of the gene located within or next to this switch. Many bacterial species can utilize inversion to change the expression of certain genes for the benefit of the bacterium during infection.[2] The inversion event can be simple by involving the toggle in expression of one gene, like E. coli pilin expression, or more complicated by involving multiple genes in the expression of multiple types of flagellin by Salmonella enterica serovar Typhimurium.[4] Fimbrial adhesion by the type I fimbriae in E. coli undergoes site specific inversion to regulate the expression of fimA, the major subunit of the pili, depending on the stage of infection. The invertible element has a promoter within it that depending on the orientation will turn on or off the transcription of fimA. The inversion is mediated by two recombinases, FimB and FimE, and regulatory proteins H-NS, Integration Host Factor (IHF) and Leucine responsive protein (LRP). The FimE recombinase has the capability to only invert the element and turn expression from on to off while FimB can mediate the inversion in both directions.[5]
Insertion-excision
If excision is precise and the original sequence of DNA is restored, reversible phase variation can be mediated by transposition. Phase variation mediated by transposition targets specific DNA sequences.[6] P. atlantica contains an eps locus that encodes extracellular polysaccharide and the ON or OFF expression of this locus is controlled by the presence or absence of IS492. Two recombinases encoded by MooV and Piv mediate the precise excision and insertion, respectively, of the insertion element IS492 in the eps locus. When IS492 is excised it becomes a circular extrachromosomal element that results in the restored expression of eps.[6][7]
Another, more complex example of site-specific DNA rearrangement is used in the flagella of Salmonella Typhimurium. In the usual phase, a promoter sequence promotes the expression of the H2 flagella gene along with a repressor of H1 flagella gene. Once this promoter sequence is inverted by the hin gene the repressor is turned off as is H2 allowing H1 to be expressed.
Gene conversion
Gene conversion is another example of a type of phase variation. Type IV pili of Neisseria gonorrhoeae are controlled in this way. There are several copies of the gene coding for these pili (the Pil gene) but only one is expressed at any given time. This is referred to as the PilE gene. The silent versions of this gene, PilS, can use homologous recombination to combine with parts of the PilE gene and thus create a different phenotype. This allows for up to 10,000,000 different phenotypes of the pili .
Epigenetic modification – methylation
Unlike other mechanisms of phase variation, epigenetic modifications do not alter DNA sequence and therefore it is the phenotype that is altered not the genotype. The integrity of the genome is intact and the change incurred by methylation alters the binding of transcription factors. The outcome is the regulation of transcription resulting in switches in gene expression.[3][6] An outer membrane protein Antigen 43 (Ag43) in E. coli is controlled by phase variation mediated by two proteins, DNA-methylating enzyme deoxyadenosine methyltransferase (Dam) and the oxidative stress regulator OxyR. Ag43, located on the cell surface, is encoded by the Agn43 gene (previously designated as flu) and is important for biofilms and infection. The expression of Agn43 is dependent on the binding of the regulator protein OxyR. When OxyR is bound to the regulatory region of Agn43, which overlaps with the promoter, it inhibits transcription. The ON phase of transcription is dependent upon Dam methylating the GATC sequences in the beginning of the Agn43 gene (which happens to overlap with the OxyR binding site). When the Dam methylates the GATC sites it inhibits the OxyR from binding, allowing transcription of Ag43.[8]
Nested DNA inversion
In this form of phase variation. The promoter region of the genome can move from one copy of a gene to another through homologous recombination. This occurs with Campylobacter fetus surface proteins. The several different surface antigen proteins are all silent apart from one and all share a conserved region at the 5' end. The promoter sequence can then move between these conserved regions and allow expression of a different gene .
Slipped strand mispairing
Slipped strand mispairing (SSM) is a process that produces mispairing of short repeat sequences between the mother and daughter strand during DNA synthesis.[2] This RecA-independent mechanism can transpire during either DNA replication or DNA repair and can be on the leading or lagging strand. SSM can result in an increase or decrease in the number of short repeat sequences. The short repeat sequences are 1 to 7 nucleotides and can be homogeneous or heterogeneous repetitive DNA sequences.[4]
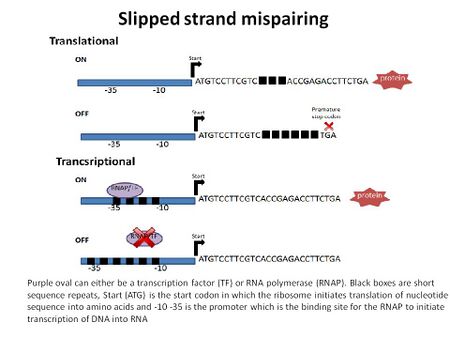
Altered gene expression is a result of SSM and depending where the increase or decrease of the short repeat sequences occurs in relation to the promoter will either regulate at the level of transcription or translation.[9] The outcome is an ON or OFF phase of a gene or genes.
Transcriptional regulation (bottom portion of figure) occurs in several ways. One possible way is if the repeats are located in the promoter region at the RNA polymerase binding site, -10 and -35 upstream of the gene(s). The opportunistic pathogen H. influenzae has two divergently oriented promoters and fimbriae genes hifA and hifB. The overlapping promoter regions have repeats of the dinucleotide TA in the -10 and -35 sequences. Through SSM the TA repeat region can undergo addition or subtraction of TA dinucleotides which results in the reversible ON phase or OFF phase of transcription of the hifA and hifB.[4][10] The second way that SSM induces transcriptional regulation is by changing the short repeat sequences located outside the promoter. If there is a change in the short repeat sequence it can affect the binding of a regulatory protein, such as an activator or repressor. It can also lead to differences in post-transcriptional stability of mRNA.[6]
Translation of a protein can be regulated by SSM if the short repeat sequences are in the coding region of the gene (top portion of the figure). Changing the number of repeats in the open reading frame can affect the codon sequence by adding a premature stop codon or by changing the sequence of the protein. This often results in a truncated (in the case of a premature stop codon) and/or nonfunctional protein.
References
- ↑ "Microbial Primer: Phase variation - survival and adaptability by generation of a diverse population". Microbiology 170 (9): 001492. September 2024. doi:10.1099/mic.0.001492. PMID 39222353.
- ↑ 2.0 2.1 2.2 2.3 "Molecular switches--the ON and OFF of bacterial phase variation". Mol Microbiol 33 (5): 919–32. 1999. doi:10.1046/j.1365-2958.1999.01555.x. PMID 10476027.
- ↑ 3.0 3.1 Bayliss CD (2009). "Determinants of phase variation rate and the fitness implications of differing rates for bacterial pathogens and commensals". FEMS Microbiol Rev 33 (3): 504–520. doi:10.1111/j.1574-6976.2009.00162.x. PMID 19222587.
- ↑ 4.0 4.1 4.2 "Phase and antigenic variation mediated by genome modifications". Antonie van Leeuwenhoek 94 (4): 493–515. 2008. doi:10.1007/s10482-008-9267-6. PMID 18663597.
- ↑ "Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media". J Bacteriol 175 (19): 6186–93. 1993. doi:10.1128/jb.175.19.6186-6193.1993. PMID 8104927.
- ↑ 6.0 6.1 6.2 6.3 "Phase and antigenic variation in bacteria". Clin Microbiol Rev 17 (3): 581–611. 2004. doi:10.1128/CMR.17.3.581-611.2004. PMID 15258095.
- ↑ "Chromosomal context directs high-frequency precise excision of IS492 in Pseudoalteromonas atlantica". Proc Natl Acad Sci USA 104 (6): 1901–1906. 2007. doi:10.1073/pnas.0608633104. PMID 17264213. Bibcode: 2007PNAS..104.1901H.
- ↑ "Regulation and function of Ag43 (flu)". Annu Rev Microbiol 62: 153–169. 2008. doi:10.1146/annurev.micro.62.081307.162938. PMID 18785838.
- ↑ "Slipped strand mispairing can function as a phase variation mechanism in Escherichia coli". J Bacteriol 185 (23): 6990–6994. 2003. doi:10.1128/JB.185.23.6990-6994.2003. PMID 14617664.
- ↑ "Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region". Cell 73 (6): 1187–96. 1993. doi:10.1016/0092-8674(93)90647-9. PMID 8513502.
 |
