Biology:Protospacer adjacent motif
A protospacer adjacent motif (PAM) is a 2–6-base pair DNA sequence immediately following the DNA sequence targeted by the Cas9 nuclease in the CRISPR bacterial adaptive immune system.[1] The PAM is a component of the invading virus or plasmid, but is not found in the bacterial host genome and hence is not a component of the bacterial CRISPR locus. Cas9 will not successfully bind to or cleave the target DNA sequence if it is not followed by the PAM sequence.[2][3][4][5] PAM is an essential targeting component which distinguishes bacterial self from non-self DNA, thereby preventing the CRISPR locus from being targeted and destroyed by the CRISPR-associated nuclease.[6]
Spacers/protospacers
In a bacterial genome, CRISPR loci contain "spacers" (viral DNA inserted into a CRISPR locus) that in type II adaptive immune systems were created from invading viral or plasmid DNA (called "protospacers"). Upon subsequent invasion, a CRISPR-associated nuclease such as Cas9 attaches to a tracrRNA–crRNA complex, which guides Cas9 to the invading protospacer sequence. But Cas9 will not cleave the protospacer sequence unless there is an adjacent PAM sequence. The spacer in the bacterial CRISPR loci will not contain a PAM sequence, and thus will not be cut by the nuclease, but the protospacer in the invading virus or plasmid will contain the PAM sequence, and thus will be cleaved by the Cas9 nuclease.[4] In genome editing applications, a short oligonucleotide known as a guide RNA (gRNA) is synthesized to perform the function of the tracrRNA–crRNA complex in recognizing gene sequences having a PAM sequence at the 3'-end, thereby "guiding" the nuclease to a specific sequence which the nuclease is capable of cutting.[7][8]
PAM sequences
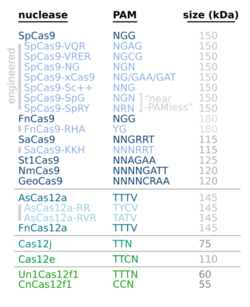
The canonical PAM is the sequence 5'-NGG-3', where "N" is any nucleobase followed by two guanine ("G") nucleobases.[9] Guide RNAs can transport Cas9 to any locus in the genome for gene editing, but no editing can occur at any site other than one at which Cas9 recognizes PAM. The canonical PAM is associated with the Cas9 nuclease of Streptococcus pyogenes (designated SpCas9), whereas different PAMs are associated with the Cas9 proteins of the bacteria Neisseria meningitidis, Treponema denticola, and Streptococcus thermophilus.[10] 5'-NGA-3' can be a highly efficient non-canonical PAM for human cells, but efficiency varies with genome location.[11] Attempts have been made to engineer Cas9s to recognize different PAMs in order to improve the ability of CRISPR-Cas9 to edit genes at any desired genome location.[12]
The Cas9 of Francisella novicida recognizes the canonical PAM sequence 5'-NGG-3', but has been engineered to recognize 5'-YG-3' (where "Y" is a pyrimidine[13]), thus adding to the range of possible Cas9 targets.[14] The Cpf1 nuclease of Francisella novicida recognizes the PAM 5'-TTTN-3'[15] or 5'-YTN-3'.[16]
Aside from CRISPR-Cas9 and CRISPR-Cpf1, there are doubtless many yet undiscovered nucleases and PAMs.[17]
CRISPR/Cas13a (formerly C2c2[18]) from the bacterium Leptotrichia shahii is an RNA-guided CRISPR system that targets sequences in RNA rather than DNA. PAM is not relevant for an RNA-targeting CRISPR, although a guanine flanking the target negatively affects efficacy, and has been designated a "protospacer flanking site" (PFS).[19]
GUIDE-Seq
A technology called GUIDE-Seq has been devised to assay off-target cleavages produced by such gene editing.[20] The PAM requirement can be exploited to specifically target single-nucleotide heterozygous mutations while exerting no aberrant effects on wild-type alleles.[21]
See also
- CRISPR
- CRISPR/Cpf1
External links
References
- ↑ "Protospacer recognition motifs: mixed identities and functional diversity". RNA Biology 10 (5): 891–899. 2013. doi:10.4161/rna.23764. PMID 23403393.
- ↑ "Short motif sequences determine the targets of the prokaryotic CRISPR defence system". Microbiology 155 (Pt 3): 733–740. 2009. doi:10.1099/mic.0.023960-0. PMID 19246744. http://mic.sgmjournals.org/content/155/3/733.long.
- ↑ "Protospacer recognition motifs: mixed identities and functional diversity". RNA Biology 10 (5): 891–899. 2013. doi:10.4161/rna.23764. PMID 23403393. PMC 3737346. https://www.landesbioscience.com/journals/rnabiology/article/23764/.
- ↑ 4.0 4.1 "A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity". Science 337 (6096): 816–821. 2012. doi:10.1126/science.1225829. PMID 22745249. Bibcode: 2012Sci...337..816J.
- ↑ "DNA interrogation by the CRISPR RNA-guided endonuclease Cas9". Nature 507 (7490): 62–67. 2014. doi:10.1038/nature13011. PMID 24476820. Bibcode: 2014Natur.507...62S.
- ↑ "Cas9 as a versatile tool for engineering biology". Nature Methods 10 (10): 957–963. 2013. doi:10.1038/nmeth.2649. PMID 24076990.
- ↑ "RNA-guided human genome engineering via Cas9". Science 339 (6121): 823–826. 2013. doi:10.1126/science.1232033. PMID 23287722. Bibcode: 2013Sci...339..823M.
- ↑ "Multiplex genome engineering using CRISPR/Cas systems". Science 339 (6121): 819–823. 2013. doi:10.1126/science.1231143. PMID 23287718. Bibcode: 2013Sci...339..819C.
- ↑ "Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease". Nature 513 (7519): 569–573. 2014. doi:10.1038/nature13579. PMID 25079318. Bibcode: 2014Natur.513..569A.
- ↑ "Orthogonal Cas9 proteins for RNA-guided gene regulation and editing". Nature Methods 10 (11): 1116–1123. 2013. doi:10.1038/nmeth.2681. PMID 24076762.
- ↑ "Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells". Scientific Reports 4: 5405. 2014. doi:10.1038/srep05405. PMID 24956376. Bibcode: 2014NatSR...4E5405Z.
- ↑ "Engineered CRISPR-Cas9 nucleases with altered PAM specificities". Nature 523 (7561): 481–485. 2015. doi:10.1038/nature14592. PMID 26098369. Bibcode: 2015Natur.523..481K.
- ↑ "Nucleotide Codes, Amino Acid Codes, and Genetic Codes". KEGG: Kyoto Encyclopedia of Genes and Genomes. July 15, 2014. http://www.genome.jp/kegg/catalog/codes1.html.
- ↑ "Structure and Engineering of Francisella novicida Cas9". Cell 164 (5): 950–961. 2016. doi:10.1016/j.cell.2016.01.039. PMID 26875867.
- ↑ "Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system". Cell 163 (3): 759–771. 2015. doi:10.1016/j.cell.2015.09.038. PMID 26422227.
- ↑ "The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA". Nature 532 (7600): 517–521. 2016. doi:10.1038/nature17945. PMID 27096362. Bibcode: 2016Natur.532..517F.
- ↑ "Even CRISPR". The Economist. ISSN 0013-0613. https://www.economist.com/news/science-and-technology/21668031-scientists-have-found-yet-another-way-edit-genomes-suggesting-such-technology-will.
- ↑ "Diversity and evolution of class 2 CRISPR-Cas systems". Nat. Rev. Microbiol. 15 (3): 169–182. 2017. doi:10.1038/nrmicro.2016.184. PMID 28111461.
- ↑ "C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector". Science 353 (6299): aaf5573. 2016. doi:10.1126/science.aaf5573. PMID 27256883.
- ↑ "GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleas". Nature Biotechnology 33 (2): 187–197. 2015. doi:10.1038/nbt.3117. PMID 25513782.
- ↑ "Exploiting the CRISPR/Cas9 PAM Constraint for Single-Nucleotide Resolution Interventions". PLoS One 11 (1): e0144970. 2016. doi:10.1371/journal.pone.0144970. PMID 26788852. Bibcode: 2016PLoSO..1144970L.
 |

