Biology:R-loop

An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA. R-loops may be formed in a variety of circumstances and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loops; the "R" in this case represents the involvement of an RNA moiety.
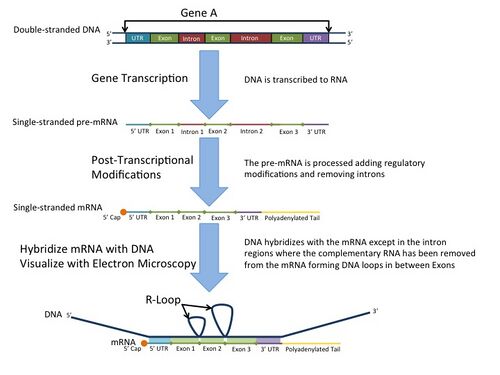
In the laboratory, R-loops may also be created by the hybridization of mature mRNA with double-stranded DNA under conditions favoring the formation of a DNA-RNA hybrid; in this case, the intron regions (which have been spliced out of the mRNA) form single-stranded DNA loops, as they cannot hybridize with complementary sequence in the mRNA.[1]
History
R-looping was first described in 1976.[2] Independent R-looping studies from the laboratories of Richard J. Roberts and Phillip A. Sharp showed that protein coding adenovirus genes contained DNA sequences that were not present in the mature mRNA.[3][4] Roberts and Sharp were awarded the Nobel Prize in 1993 for independently discovering introns. After their discovery in adenovirus, introns were found in a number of eukaryotic genes such as the eukaryotic ovalbumin gene (first by the O'Malley laboratory, then confirmed by other groups),[5][6] hexon DNA,[3] and extrachromosomal rRNA genes of Tetrahymena thermophila.[7]
In the mid-1980s, development of an antibody that binds specifically to the R-loop structure opened the door for immunofluorescence studies, as well as genome-wide characterization of R-loop formation by DRIP-seq.[8]
R-loop mapping
R-loop mapping is a laboratory technique used to distinguish introns from exons in double-stranded DNA.[9] These R-loops are visualized by electron microscopy and reveal intron regions of DNA by creating unbound loops at these regions.[10]
R-loops in vivo
The potential for R-loops to serve as replication primers was demonstrated in 1980.[11] In 1994, R-loops were demonstrated to be present in vivo through analysis of plasmids isolated from E. coli mutants carrying mutations in topoisomerase.[12] This discovery of endogenous R-loops, in conjunction with rapid advances in genetic sequencing technologies, inspired a blossoming of R-loop research in the early 2000s that continues to this day.[13]
Regulation of R-loop formation and resolution
RNaseH enzymes are the primary proteins responsible for the dissolution of R-loops, acting to degrade the RNA moiety in order to allow the two complementary DNA strands to anneal.[14] Research over the past decade has identified more than 50 proteins that appear to influence R-loop accumulation, and while many of them are believed to contribute by sequestering or processing newly transcribed RNA to prevent re-annealing to the template, mechanisms of R-loop interaction for many of these proteins remain to be determined.[15]
Roles of R-loops in genetic regulation
R-loop formation is a key step in immunoglobulin class switching, a process that allows activated B cells to modulate antibody production.[16] They also appear to play a role in protecting some active promoters from methylation.[17] The presence of R-loops can also inhibit transcription.[18] Additionally, R-loop formation appears to be associated with “open” chromatin, characteristic of actively transcribed regions.[19][20]
R-loops as genetic damage
When unscheduled R-loops form, they can cause damage by a number of different mechanisms.[21] Exposed single-stranded DNA can come under attack by endogenous mutagens, including DNA-modifying enzymes such as activation-induced cytidine deaminase, and can block replication forks to induce fork collapse and subsequent double-strand breaks.[22] As well, R-loops may induce unscheduled replication by acting as a primer.[11][20]
R-loop accumulation has been associated with a number of diseases, including amyotrophic lateral sclerosis type 4 (ALS4), ataxia oculomotor apraxia type 2 (AOA2), Aicardi–Goutières syndrome, Angelman syndrome, Prader–Willi syndrome, and cancer.[13]
R-loops, Introns and DNA damage
Introns are non-coding regions within genes that are transcribed along with the coding regions of genes, but are subsequently removed from the primary RNA transcript by splicing. Actively transcribed regions of DNA often form R-loops that are vulnerable to DNA damage. Introns reduce R-loop formation and DNA damage in highly expressed yeast genes.[23] Genome-wide analysis showed that intron-containing genes display decreased R-loop levels and decreased DNA damage compared to intron-less genes of similar expression in both yeast and humans.[23] Inserting an intron within an R-loop prone gene can also suppress R-loop formation and recombination. Bonnet et al. (2017)[23] speculated that the function of introns in maintaining genetic stability may explain their evolutionary maintenance at certain locations, particularly in highly expressed genes.
See also
References
- ↑ Wang, Kang; Wang, Honghong; Li, Conghui; Yin, Zhinang; Xiao, Ruijing; Li, Qiuzi; Xiang, Ying; Wang, Wen et al. (2021-02-19). "Genomic profiling of native R loops with a DNA-RNA hybrid recognition sensor" (in en). Science Advances 7 (8). doi:10.1126/sciadv.abe3516. ISSN 2375-2548. PMID 33597247.
- ↑ "Hybridization of RNA to double-stranded DNA: formation of R-loops". Proceedings of the National Academy of Sciences of the United States of America 73 (7): 2294–8. July 1976. doi:10.1073/pnas.73.7.2294. PMID 781674. Bibcode: 1976PNAS...73.2294T.
- ↑ 3.0 3.1 "Spliced segments at the 5' terminus of adenovirus 2 late mRNA". Proceedings of the National Academy of Sciences of the United States of America 74 (8): 3171–5. August 1977. doi:10.1073/pnas.74.8.3171. PMID 269380. Bibcode: 1977PNAS...74.3171B.
- ↑ "An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA". Cell 12 (1): 1–8. September 1977. doi:10.1016/0092-8674(77)90180-5. PMID 902310.
- ↑ "The ovalbumin gene: structural sequences in native chicken DNA are not contiguous". Proceedings of the National Academy of Sciences of the United States of America 75 (5): 2205–9. May 1978. doi:10.1073/pnas.75.5.2205. PMID 276861. Bibcode: 1978PNAS...75.2205L.
- ↑ "No more than seven interruptions in the ovalbumin gene: comparison of genomic and double-stranded cDNA sequences". Nucleic Acids Research 7 (2): 321–34. September 1979. doi:10.1093/nar/7.2.321. PMID 493147.
- ↑ "Localization of transcribed regions on extrachromosomal ribosomal RNA genes of Tetrahymena thermophila by R-loop mapping". Proceedings of the National Academy of Sciences of the United States of America 76 (10): 5051–5. October 1979. doi:10.1073/pnas.76.10.5051. PMID 291921. Bibcode: 1979PNAS...76.5051C.
- ↑ "Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids". Journal of Immunological Methods 89 (1): 123–30. May 1986. doi:10.1016/0022-1759(86)90040-2. PMID 2422282.
- ↑ "The use of R-looping for structural gene identification and mRNA purification". Nucleic Acids Research 6 (7): 2483–97. June 1979. doi:10.1093/nar/6.7.2483. PMID 379820.
- ↑ King RC, Stansfield WD, Mulligan PK (2007). A Dictionary of Genetics. Oxford University Press 7.
- ↑ 11.0 11.1 "Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H". Proceedings of the National Academy of Sciences of the United States of America 77 (5): 2450–4. May 1980. doi:10.1073/pnas.77.5.2450. PMID 6156450. Bibcode: 1980PNAS...77.2450I.
- ↑ "Hypernegative supercoiling of the DNA template during transcription elongation in vitro". The Journal of Biological Chemistry 269 (3): 2068–74. January 1994. doi:10.1016/S0021-9258(17)42136-3. PMID 8294458.
- ↑ 13.0 13.1 "Out of balance: R-loops in human disease". PLOS Genetics 10 (9): e1004630. September 2014. doi:10.1371/journal.pgen.1004630. PMID 25233079.
- ↑ "Ribonuclease H: the enzymes in eukaryotes". The FEBS Journal 276 (6): 1494–505. March 2009. doi:10.1111/j.1742-4658.2009.06908.x. PMID 19228196.
- ↑ "Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip". PLOS Genetics 10 (4): e1004288. April 2014. doi:10.1371/journal.pgen.1004288. PMID 24743342.
- ↑ "Mechanism of R-loop formation at immunoglobulin class switch sequences". Molecular and Cellular Biology 28 (1): 50–60. January 2008. doi:10.1128/mcb.01251-07. PMID 17954560.
- ↑ "R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters". Molecular Cell 45 (6): 814–25. March 2012. doi:10.1016/j.molcel.2012.01.017. PMID 22387027.
- ↑ "A novel mode for transcription inhibition mediated by PNA-induced R-loops with a model in vitro system". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1861 (2): 158–166. February 2018. doi:10.1016/j.bbagrm.2017.12.008. PMID 29357316.
- ↑ "R loops are linked to histone H3 S10 phosphorylation and chromatin condensation". Molecular Cell 52 (4): 583–90. November 2013. doi:10.1016/j.molcel.2013.10.006. PMID 24211264.
- ↑ 20.0 20.1 "The Yin and Yang of R-loop biology". Current Opinion in Cell Biology 34: 39–45. June 2015. doi:10.1016/j.ceb.2015.04.008. PMID 25938907.
- ↑ "R-loop generation during transcription: Formation, processing and cellular outcomes". DNA Repair 71: 69–81. November 2018. doi:10.1016/j.dnarep.2018.08.009. PMID 30190235.
- ↑ "Breaking bad: R-loops and genome integrity". Trends in Cell Biology 25 (9): 514–22. September 2015. doi:10.1016/j.tcb.2015.05.003. PMID 26045257.
- ↑ 23.0 23.1 23.2 "Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability". Molecular Cell 67 (4): 608–621.e6. August 2017. doi:10.1016/j.molcel.2017.07.002. PMID 28757210.
 |
