Biology:RsmX
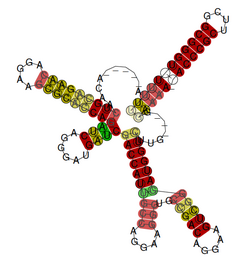
The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli,[2] Erwinia,[3] Salmonella,[4] Vibrio[5] and Pseudomonas.[6] These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs.[7] Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.[8]
In Pseudomonas spp., three rsm ncRNAs have been identified. These are RsmX (approximately 115 nt), RsmY (approximately 120 nt) and RsmZ (approximately 145 nt). Expression of all three ncRNAs is population density dependent, with maximal expression occurring at the end of exponential phase.[9] Further, expression of all three ncRNAs is dependent upon the response regulator, GacA, which activates transcription of the ncRNAs by binding a conserved upstream activating sequence (UAS) in the promoter region.[10] Typically, pseudomonads contain a single copy of each of RsmY and RsmZ, however the copy number of RsmX is more variable. For example, P. aeruginosa contains no copies of RsmX and P. syringae pathovars contain five copies.[1]
See also
- CsrB/RsmB RNA family
- CsrC RNA family
- PrrB/RsmZ RNA family
- RsmY RNA family
- CsrA protein
References
- ↑ 1.0 1.1 Moll et al. (2010) "Construction of an rsmX co-variance model and identification of five rsmX-like ncRNAs in Pseudomonas syringae pv. tomato DC3000." RNA Biology 7(5):
- ↑ "Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties". J. Bacteriol. 175 (15): 4744–4755. August 1993. doi:10.1128/jb.175.15.4744-4755.1993. PMID 8393005.
- ↑ Liu et al. (1998) "Characterization of a novel RNA regulator of Erwinia caratovora ssp. caratovora that controls production of extracellular enzymes and secondary metabolites." Molecular Microbiology 29: 219–234
- ↑ "Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium". Infect. Immun. 74 (1): 331–339. January 2006. doi:10.1128/IAI.74.1.331-339.2006. PMID 16368988.
- ↑ "CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae". Mol. Microbiol. 58 (4): 1186–1202. November 2005. doi:10.1111/j.1365-2958.2005.04902.x. PMID 16262799.
- ↑ "Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0". J. Bacteriol. 184 (4): 1046–1056. February 2002. doi:10.1128/jb.184.4.1046-1056.2002. PMID 11807065.
- ↑ "A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens". J. Biol. Chem. 279 (24): 25066–25074. June 2004. doi:10.1074/jbc.M401870200. PMID 15031281.
- ↑ "Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour". Mol. Microbiol. 67 (2): 241–253. January 2008. doi:10.1111/j.1365-2958.2007.06042.x. PMID 18047567.
- ↑ "Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0". Proc. Natl. Acad. Sci. U.S.A. 102 (47): 17136–17141. November 2005. doi:10.1073/pnas.0505673102. PMID 16286659. Bibcode: 2005PNAS..10217136K.
- ↑ "GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens". Appl. Environ. Microbiol. 76 (5): 1497–1506. March 2010. doi:10.1128/AEM.02014-09. PMID 20048056. Bibcode: 2010ApEnM..76.1497H.
 |

