Biology:Saffman–Delbrück model
The Saffman–Delbrück model describes a lipid membrane as a thin layer of viscous fluid, surrounded by a less viscous bulk liquid. This picture was originally proposed to determine the diffusion coefficient of membrane proteins, but has also been used to describe the dynamics of fluid domains within lipid membranes. The Saffman–Delbrück formula is often applied to determine the size of an object embedded in a membrane from its observed diffusion coefficient, and is characterized by the weak logarithmic dependence of diffusion constant on object radius.
Origin
In a three-dimensional highly viscous liquid, a spherical object of radius a has diffusion coefficient
- [math]\displaystyle{ D_{3D} = \frac{k_B T}{6 \pi \eta a} }[/math]
by the well-known Stokes–Einstein relation. By contrast, the diffusion coefficient of a circular object embedded in a two-dimensional fluid diverges; this is Stokes' paradox. In a real lipid membrane, the diffusion coefficient may be limited by:
- the size of the membrane
- the inertia of the membrane (finite Reynolds number)
- the effect of the liquid surrounding the membrane
Philip Saffman and Max Delbrück calculated the diffusion coefficient for these three cases, and showed that Case 3 was the relevant effect.[1]
Saffman–Delbrück formula
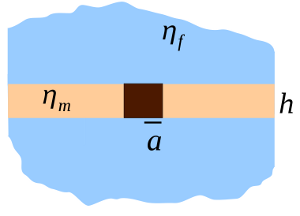
The diffusion coefficient of a cylindrical inclusion of radius [math]\displaystyle{ a }[/math] in a membrane with thickness [math]\displaystyle{ h }[/math] and viscosity [math]\displaystyle{ \eta_m }[/math], surrounded by bulk fluid with viscosity [math]\displaystyle{ \eta_f }[/math] is:
- [math]\displaystyle{ D_{sd} = \frac{k_B T}{4 \pi \eta_m h} \left[\ln(2 L_{sd} / a) - \gamma\right] }[/math]
where the Saffman–Delbrück length [math]\displaystyle{ L_{sd} = \frac{h \eta_m}{2 \eta_f} }[/math] and [math]\displaystyle{ \gamma\approx 0.577 }[/math] is the Euler–Mascheroni constant. Typical values of [math]\displaystyle{ L_{sd} }[/math] are 0.1 to 10 micrometres.[2] This result is an approximation applicable for radii [math]\displaystyle{ a \ll L_{sd} }[/math], which is appropriate for proteins ([math]\displaystyle{ a\approx }[/math] nm), but not for micrometre-scale lipid domains.
The Saffman–Delbrück formula predicts that diffusion coefficients [math]\displaystyle{ D_{sd} }[/math] will only depend weakly on the size of the embedded object; for example, if [math]\displaystyle{ L_{sd} = 1 \mu m }[/math], changing [math]\displaystyle{ a }[/math] from 1 nm to 10 nm only reduces the diffusion coefficient [math]\displaystyle{ D_{sd} }[/math] by 30%.
Beyond the Saffman–Delbrück length
Hughes, Pailthorpe, and White extended the theory of Saffman and Delbrück to inclusions with any radii [math]\displaystyle{ a }[/math];[3] for [math]\displaystyle{ a \gg L_{sd} }[/math],
- [math]\displaystyle{ D \to \frac{k_B T}{8 \eta_m h} \frac{L_{sd}}{a} = \frac{k_B T}{16 \eta_f a} }[/math]
A useful formula that produces the correct diffusion coefficients between these two limits is [2]
- [math]\displaystyle{ D = \frac{k_B T}{4 \pi \eta_m h} \left[\ln(2/\epsilon) - \gamma + 4\epsilon/\pi - (\epsilon^2/2)\ln(2/\epsilon)\right] \left[1 - (\epsilon^3/\pi) \ln(2/\epsilon) + c_1 \epsilon^{b_1} / (1 + c_2 \epsilon^{b_2}) \right]^{-1} }[/math]
where [math]\displaystyle{ \epsilon = a / L_{sd} }[/math], [math]\displaystyle{ b_1 = 2.74819 }[/math], [math]\displaystyle{ b_2 = 0.51465 }[/math], [math]\displaystyle{ c_1 = 0.73761 }[/math], and [math]\displaystyle{ c_2 = 0.52119 }[/math]. Please note that the original version of [2] has a typo in [math]\displaystyle{ b_2 }[/math]; the value in the correction[4] to that article should be used.
Experimental studies
Though the Saffman–Delbruck formula is commonly used to infer the sizes of nanometer-scale objects, recent controversial[5] experiments on proteins have suggested that the diffusion coefficient's dependence on radius [math]\displaystyle{ a }[/math] should be [math]\displaystyle{ a^{-1} }[/math] instead of [math]\displaystyle{ \ln(a) }[/math].[6] However, for larger objects (such as micrometre-scale lipid domains), the Saffman–Delbruck model (with the extensions above) is well-established [2][7][8]
Extending Saffman–Delbrück for Hydrodynamic Coupling of Proteins within Curved Lipid Bilayer Membranes
The Saffman–Delbrück approach has also been extended in recent works for modeling hydrodynamic interactions between proteins embedded within curved lipid bilayer membranes, such as in vesicles and other structures.[9][10][11][12] These works use related formulations to study the roles of the membrane hydrodynamic coupling and curvature in the collective drift-diffusion dynamics of proteins within bilayer membranes. Various models of the protein inclusions within curved membranes have been developed, including models based on series truncations,[9] immersed boundary methods,[11] and fluctuating hydrodynamics.[12]
References
- ↑ P. G. Saffman and M. Delbrück, Brownian motion in biological membranes, Proc. Natl. Acad. Sci. USA, vol. 72 p. 3111–3113 1975
- ↑ Jump up to: 2.0 2.1 2.2 2.3 Petrov, EP; Schwille, P (2008). "Translational diffusion in lipid membranes beyond the Saffman-Delbruck approximation". Biophys J 94 (5): L41-3. doi:10.1529/biophysj.107.126565. PMID 18192354. Bibcode: 2008BpJ....94L..41P.
- ↑ Hughes, B.D.; Pailthorpe, B.A.; White, L.R. (1981). "The translational and rotational drag on a cylinder moving in a membrane". J. Fluid Mech. 110: 349–372. doi:10.1017/S0022112081000785. Bibcode: 1981JFM...110..349H.
- ↑ Petrov; Schwille (Jul 2012). "Correction: Translational Diffusion in Lipid Membranes beyond the Saffman-Delbrück Approximation". Biophys. J. 103 (2): 375. doi:10.1016/j.bpj.2012.06.032. Bibcode: 2012BpJ...103..375P.
- ↑ Weiß (2013). "Quantifying the Diffusion of Membrane Proteins and Peptides in Black Lipid Membranes with 2-Focus Fluorescence Correlation Spectroscopy". Biophys. J. 105 (2): 455–462. doi:10.1016/j.bpj.2013.06.004. PMID 23870266. Bibcode: 2013BpJ...105..455W.
- ↑ Gambin, Y. (2006). "Lateral mobility of proteins in liquid membranes revisited". Proc. Natl. Acad. Sci. USA 103 (7): 2098–2102. doi:10.1073/pnas.0511026103. PMID 16461891. Bibcode: 2006PNAS..103.2098G.
- ↑ Klingler, J.F.; McConnell, H.M. (1993). "Brownian motion and fluid mechanics of lipid monolayer domains". J. Phys. Chem. 93 (22): 6096–6100. doi:10.1021/j100124a052.
- ↑ Cicuta, P.; Veatch, S.L.; Keller, S.L. (2007). "Diffusion of Liquid Domains in Lipid Bilayer Membranes". J. Phys. Chem. B 111 (13): 3328–3331. doi:10.1021/jp0702088. PMID 17388499.
- ↑ Jump up to: 9.0 9.1 Henle, Mark L.; Levine, Alex J. (12 January 2010). "Hydrodynamics in curved membranes: The effect of geometry on particulate mobility". Physical Review E 81 (1): 011905. doi:10.1103/PhysRevE.81.011905.
- ↑ Daniels, D. R. (October 2016). "Curvature correction to the mobility of fluid membrane inclusions". The European Physical Journal E 39 (10): 96. doi:10.1140/epje/i2016-16096-3.
- ↑ Jump up to: 11.0 11.1 Sigurdsson, Jon Karl; Atzberger, Paul J. (2016). "Hydrodynamic coupling of particle inclusions embedded in curved lipid bilayer membranes". Soft Matter 12 (32): 6685–6707. doi:10.1039/C6SM00194G.
- ↑ Jump up to: 12.0 12.1 Rower, David A.; Padidar, Misha; Atzberger, Paul J. (15 April 2022). "Surface fluctuating hydrodynamics methods for the drift-diffusion dynamics of particles and microstructures within curved fluid interfaces". Journal of Computational Physics 455: 110994. doi:10.1016/j.jcp.2022.110994.
 |