Biology:skipping-rope RNA motif
| skipping-rope | |
|---|---|
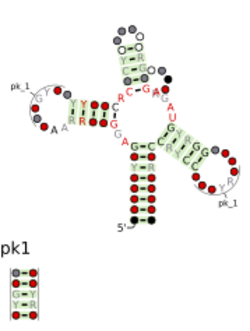 Consensus secondary structure and sequence conservation of skipping-rope RNA | |
| Identifiers | |
| Symbol | skipping-rope |
| Rfam | RF02924 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0005836 |
| PDB structures | PDBe |
The skipping-rope RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] skipping-rope motif RNAs are found in multiple phyla: Bacillota, Fusobacteriota, Pseudomonadota and Spirochaetota. A skipping-rope RNA was also found in a purified phage, specifically the phage Bacillus phage SPbeta, which infects Bacillus organisms that fit into the phylum Bacillota. Therefore, skipping-rope RNAs likely function, at least sometimes, to perform a function useful to phages.
skipping-rope RNAs likely function in trans as small RNAs, and are often immediately followed on their 3′ ends by Rho-independent transcription terminators. Genes that encode apparently homologous proteins are often located nearby to skipping-rope RNAs. These genes can occur 5′ or 3′ relative to the RNA, and on the same or opposite DNA strand. Occasionally, these proteins match the DUF3800 conserved protein domain,[1] and so skipping-rope RNAs might be an example of DUF3800 RNA motifs. These properties are also similar to the Drum RNA motif.
References
- ↑ 1.0 1.1 "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
 |

