Biology:Succinate dehydrogenase subunit E
| SdhE | |||||||||
|---|---|---|---|---|---|---|---|---|---|
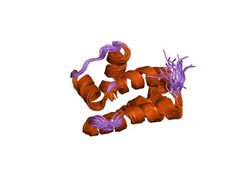 Solution NMR structure of protein NMA1147 from Neisseria meningitidis. Northeast structural genomics consortium target mr19 | |||||||||
| Identifiers | |||||||||
| Symbol | SdhE | ||||||||
| Pfam | PF03937 | ||||||||
| InterPro | IPR005631 | ||||||||
| |||||||||
In molecular biology, the protein domain named Sdh5 is also named SdhE which stands for succinate dehydrogenase protein E. In the past, it has also been named YgfY and DUF339.[1] Another name for sdhE is succinate dehydrogenase assembly factor 2 (sdhaf2).[2] This protein belongs to a group of highly conserved small proteins found in both eukaryotes and prokaryotes, including NMA1147 from Neisseria meningitidis [3] and YgfY from Escherichia coli.[4] The SdhE protein is found on the mitochondrial membrane is it is important for creating energy via a process named the electron transport chain.[1]
Function
The function of SDHE has been described as a flavinator of succinate dehydrogenase. SdhE works as a co-factor chaperone that incorporates FAD into SdhA. This results in SdhA flavinylation which is required for the proper function succinate dehydrogenase. More recent scientific studies indicate that SdhE is required by bacteria in order to grow on succinate, using succinate as its only source of carbon and additionally for the function, of succinate dehydrogenase, a vital component of the electron transport chain which produces energy.[1]
Structure
The structure of these proteins consists of a complex bundle of five alpha-helices, which is composed of an up-down 3-helix bundle plus an orthogonal 2-helix bundle.[4]
Protein interactions
SdhE interacts with the catalytic subunit of the succinate dehydrogenase (SDH) complex.[5]
Human disease
The human gene named SDH5, encodes for the SdhE protein. The gene itself is located in the chromosomal position 11q13.1. Loss-of-function mutations result in paraganglioma, a neuroendocrine tumour.[5]
History
The recent studies which suggest SdhE is required for bacterial flavinylation contradict previous thoughts on SdhE. It was originally proposed that FAD incorporation into bacterial flavoproteins was an autocatalytic process. Recent studies now argue that SdhE is the first protein to be identified as required for flavinylation in bacteria. Historically, the SdhE protein was once considered a hypothetical protein.[1] YgfY was also thought to be involved in transcriptional regulation.[4]
References
- ↑ 1.0 1.1 1.2 1.3 "SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria.". J Biol Chem 287 (22): 18418–28. 2012. doi:10.1074/jbc.M111.293803. PMID 22474332.
- ↑ https://www.genecards.org/cgi-bin/carddisp.pl?gene=SDHAF2
- ↑ "NMR structure of the hypothetical protein NMA1147 from Neisseria meningitidis reveals a distinct 5-helix bundle". Proteins 55 (3): 756–8. May 2004. doi:10.1002/prot.20009. PMID 15103637.
- ↑ 4.0 4.1 4.2 "Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation". Proteins 58 (3): 759–63. February 2005. doi:10.1002/prot.20337. PMID 15593094.
- ↑ 5.0 5.1 "SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma.". Science 325 (5944): 1139–42. 2009. doi:10.1126/science.1175689. PMID 19628817.

