Biology:Tcr-seq
TCR-Seq (T-cell Receptor Sequencing) is a method used to identify and track specific T cells and their clones...[1] TCR-Seq utilizes the unique nature of a T-cell receptor (TCR) as a ready-made molecular barcode.[1] This technology can apply to both single cell sequencing technologies and high throughput screens[1]
Background
T-cell Receptor (TCR)
T cells are a part of the adaptive immune system and play a critical role in protecting the body from foreign pathogens.[2] T-cell receptors (TCRs) are a group of membrane proteins found on the surface of T cells which can bind to foreign antigens.[3] TCRs interact with major histocompatibility complexes (MHC) on cell surfaces to recognize antigens. They are heterodimers made up of predominantly α and β chains (or more rarely δ and γ chains)[4] and consist of a variable region and a constant region. Variable regions are produced through a process called VDJ recombination, which results in unique amino acid sequences for α, β, and γ chains. The result is that each TCR is unique and recognizes a specific antigen[4]
Complementarity Determining Regions (CDRs)

Complementarity determining regions (CDRs) are a part of the TCR and play an essential role in TCR-MHC interactions. CDR1 and CDR2 are encoded by V genes, while CDR3 is made from the region between V and J genes or between D and J genes (termed "VDJ genes" when referred to together).[4] CDR3 is the most variable of the CDRs, and is in direct contact with the antigen.[4][5] As such, CDR3 is used as the “barcode region” to identify unique T cell populations, as it is highly unlikely for two T cells to have the same CDR3 sequence unless they came from the same parental T cell.[4][5]
Clonality

VDJ recombination produces such a vast amount of unique TCRs that many receptors never encounter the antigen they are best suited for. When a foreign antigen is present in the body, the few T cells that recognize that antigen are positively selected for so that the body has an adequate number of T cells to mount an effective immune response.[6] The selected T cells rapidly divide and differentiate into effector T-cells through a process called clonal expansion,[7] which retains the TCR sequence (including the CDR3 sequence) that originally recognized the antigen[8]
TCR-Seq uses the unique nature of the TCR - in particular CDR3 - as a molecular barcode to track T cells through a variety of processes like differentiation and proliferation,[9] which can be used for a wide variety of purposes.
Methods
Bulk vs Single-Cell Sequencing
TCR sequencing can be performed in on pooled cell populations (“bulk sequencing”) or single cells (“single cell sequencing”).[4] Bulk sequencing is useful to explore entire TCR repertoires - all the TCRs within an individual or a sample - and to generate comparisons between repertoires of different individuals.[4] This method can sequence millions of cells in a single experiment.[5] However, one major disadvantage is that bulk sequencing cannot determine which TCR chains pair together, only the frequency within the repertoire. The large amount of TCRs sampled also means that lower abundance TCRs may not be detected[5] Single cell sequencing can determine TCR chain pairs, making them more useful for identifying specific TCRs.[4][5] Some major disadvantages of this technique are its high costs,[4] limited capacity of a few thousand cells,[5] and the necessity of live cells which may be more challenging to obtain[4]
Target Sequences
Any TCR chain can be sequenced, although the α and β chain are more commonly chosen due to their abundance in the T cell population.[4] In particular, the β chain is of interest due to its higher diversity and specificity compared to other chains. The presence of a D gene component in the β chain which is not present in the α chain allows more diverse combinations.[4][10] As well, β chains are unique to each T cell, which can be used to identify distinct T cell populations within a sample[4][5][10]
To perform TCR-sequencing, polymerase chain reaction (PCR) amplification is performed on the CDR3 region as a measure of unique T cells within a population.[4][10] The CDR3 region is chosen over CDR1 and CDR2 as it is directly responsible for antigen interactions[4][10] and is generally unique to TCRs from the same lineage, which allows identification of distinct populations[10]
Library Preparation
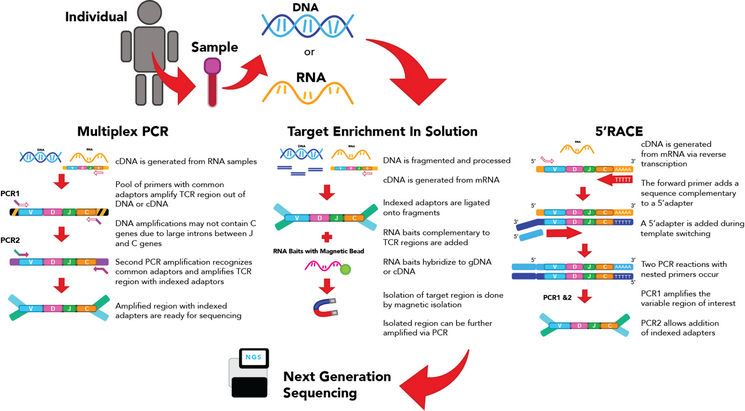
The goal of this step is to generate a library of transcripts to be sequenced. There are 3 major ways of generating a library for TCR sequencing.
Multiplex DNA
Multiplex PCR can be employed on both genomic DNA (gDNA) or RNA which has been converted to double-stranded complementary DNA (cDNA).[4] Primer pools with primer pairs targeting J and V alleles are used to amplify the CDR3 region of the TCR transcript.[4][10] The transcript goes through two or more rounds of PCR to amplify the region of interest, then adaptors are ligated onto either end of the resulting transcript.[10] This method is among the most used in the generation of libraries for TCR-seq as it can capture a great deal of the diversity of the TCR through the primer pool.[4][10] However, as it is near-impossible to optimize PCR conditions for all the primers in the pool, multiplex DNA can result in amplification bias where some CDR3 regions with primers that bind poorly may not be amplified.[5][10] This means the abundance of amplified segments may not correspond with the actual abundance within the cell[5][10]
Target Enrichment In-Solution
This method can use genomic gDNA or RNA converted to cDNA.[4][10] The starting material is first processed to generate DNA or cDNA transcripts with indexed adaptors on the 5’ and 3’ ends.[4] These transcripts are then incubated with RNA baits designed to bind to regions of interest, which is generally the CDR3 region.[4] These baits, which are normally bound to magnetic beads, can be isolated using a magnet. This allows the isolation of transcripts of the CDR3 region which can then amplified using PCR.[4] Target enrichment using RNA baits requires fewer PCR amplification steps, which may decrease amplification bias.[4] However, the efficiency of the capture by magnets may affect the diversity of the amplified transcripts.
5’-RACE
Rapid Amplification of cDNA Ends (RACE) is a method that uses RNA transcripts for generation of the library.[4][10][11] Although RACE can be applied with the 3' or the 5' end, the 5' end more commonly used for TCR-seq.[4] This method revolves around the addition of a common 5' adaptor sequence to the transcript, which can be a done a few different ways.[11][12] One method is to add on the adapter following reverse transcription. During the generation of the reverse DNA strand from the RNA template, a forward primer adds a sequence complementary to the 5'adapter, leading to template switching[4][10][11] This allows a 5' adapter to be incorporated into the cDNA when the complementary sequence is generated.[4][10][11][12] Primers can be designed to amplify the entire region from the adaptor to the constant region,[4][10][11] then adaptor ligation can be performed in a second PCR reaction. As all the different transcripts now share an identical adapter, they can be amplified using a single primer pair. As such, this method decreases amplification bias and improves the ability to detect more uncommon TCR populations with greater certainty.[4][10][11] However, as TCR transcription levels differ between cells, this method cannot provide an accurate measurement of the number of different T cell types in the sample based on the level of RNA transcripts alone[4]
Sequencing
Following generation of the library, the products can be sequenced, generally via Next Generation Sequencing (NGS).[4][10][11] Usage of machines capable of longer reads and maintains read quality at the 3’end is important, as the CDR3 region is at the 3’end of an approximately 500 base pair transcript[10]
The error rate of NGS presents a challenge for analysis of TCR repertoires.[4][10][11] Small variations in the TCR can change their specificity towards antigens,[10] and as such may be interest to researchers. However, errors in sequencing can generate a minor change that may be interpreted as a low-frequency, distinct TCR population,[10] which is a problem when analyzing changes in TCR repertoires. Efforts have been made to establish thresholds to remove low abundance reads from analysis, as well as to develop algorithms to correct these errors[10]
Applications
Generally, the data collected from TCR-seq is used to compare TCR repertoires, either between the same patient at different timepoints, or between different patients.[4][10][11] Recent studies examined the characteristics of a healthy repertoire, and found a high degree of variation in TCR β chain levels and types, though a subset is shared across different individuals.[10] However, this diversity has yet to be shown to strongly correlate with any conditions of interest, such as rates of infection or chance of cancer relapse,[10] suggesting further research is necessary.
Infectious Diseases
Clonal expansion of T cells allow the immune system to deal with a variety of infection disease with high specificity.[13] Thus, understanding changes that occur to the T cell repertoire following disease infection can early diagnosis, disease monitoring, and therapeutic development[5]
Acquired Immunodeficiency Syndrome (AIDS) is a devastating disease caused by Human Immunodeficiency Virus (HIV) infection, which results in the death of CD4+ T cells.[14] and dysfunctional CD8+ T cells.[5] Recent studies have suggested that increased TCR diversity may decrease HIV diversity and limit disease progression.[5][15] Sequencing of the TCR would also increase understanding of the progression of AIDS and predict morbidity.[5] Additionally, sequencing the TCR repertoire of individuals with natural defense against AIDs infection[16] could help development of a vaccine to limit further spread of the disease[5]
Cancer
Cancer is the uncontrolled proliferation of malignant cells which can spread throughout the body.[17] This is caused by mutations within the cancer cell, which often leads to expression of mutant proteins termed neoantigens.[5][17] Identification of these neoantigens has great therapeutic benefit, as they can be exploited to target cancer cells without harming normal cells. As CD8+ T cells can recognize some neoantigens in their TCR, sequencing of TCR repertoires can help identify potential cancer biomarkers.[5] In addition to biomarker identification, sequencing of the TCR repertoire can also track changes in cancer progression, assess responses to immunotherapy, and evaluate the tumour microenvironment for conditions that may make it permissible to cancer growth[5]
See also
References
- ↑ 1.0 1.1 1.2 Pai, Joy A.; Satpathy, Ansuman T. (August 2021). "High-throughput and single-cell T cell receptor sequencing technologies". Nature Methods 18 (8): 881–892. doi:10.1038/s41592-021-01201-8. ISSN 1548-7105. PMID 34282327.
- ↑ Marshall, Jean S.; Warrington, Richard; Watson, Wade; Kim, Harold L. (2018-09-12). "An introduction to immunology and immunopathology". Allergy, Asthma & Clinical Immunology 14 (2): 49. doi:10.1186/s13223-018-0278-1. ISSN 1710-1492. PMID 30263032.
- ↑ "T cell receptor" (in en). 2011-02-02. https://www.cancer.gov/publications/dictionaries/cancer-terms/def/t-cell-receptor.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 4.19 4.20 4.21 4.22 4.23 4.24 4.25 4.26 4.27 4.28 4.29 4.30 4.31 4.32 Rosati, Elisa; Dowds, C. Marie; Liaskou, Evaggelia; Henriksen, Eva Kristine Klemsdal; Karlsen, Tom H.; Franke, Andre (2017-07-10). "Overview of methodologies for T-cell receptor repertoire analysis". BMC Biotechnology 17 (1): 61. doi:10.1186/s12896-017-0379-9. ISSN 1472-6750. PMID 28693542.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 5.16 Mazzotti, Lucia; Gaimari, Anna; Bravaccini, Sara; Maltoni, Roberta; Cerchione, Claudio; Juan, Manel; Navarro, Europa Azucena-Gonzalez; Pasetto, Anna et al. (2022-08-02). "T-Cell Receptor Repertoire Sequencing and Its Applications: Focus on Infectious Diseases and Cancer". International Journal of Molecular Sciences 23 (15): 8590. doi:10.3390/ijms23158590. ISSN 1422-0067. PMID 35955721.
- ↑ Abbott, Joel D.; Ball, Gene; Boumpas, Dimitrios et al., eds. (2004), "Clonal expansion" (in en), Rheumatology and Immunology Therapy (Berlin, Heidelberg: Springer): pp. 226–227, doi:10.1007/3-540-29662-x_700, ISBN 978-3-540-29662-1, https://doi.org/10.1007/3-540-29662-X_700, retrieved 2023-02-28
- ↑ Kumar, Brahma V.; Connors, Thomas; Farber, Donna L. (2018-02-20). "Human T cell development, localization, and function throughout life". Immunity 48 (2): 202–213. doi:10.1016/j.immuni.2018.01.007. ISSN 1074-7613. PMID 29466753.
- ↑ Huang, Huang; Sikora, Michael J.; Islam, Saiful; Chowdhury, Roshni Roy; Chien, Yueh-hsiu; Scriba, Thomas J.; Davis, Mark M.; Steinmetz, Lars M. (2019-04-30). "Select sequencing of clonally expanded CD8 + T cells reveals limits to clonal expansion" (in en). Proceedings of the National Academy of Sciences 116 (18): 8995–9001. doi:10.1073/pnas.1902649116. ISSN 0027-8424. PMID 30992377. Bibcode: 2019PNAS..116.8995H.
- ↑ Pauken, Kristen E.; Lagattuta, Kaitlyn A.; Lu, Benjamin Y.; Lucca, Liliana E.; Daud, Adil I.; Hafler, David A.; Kluger, Harriet M.; Raychaudhuri, Soumya et al. (2022-03-01). "TCR-sequencing in cancer and autoimmunity: barcodes and beyond" (in en). Trends in Immunology 43 (3): 180–194. doi:10.1016/j.it.2022.01.002. ISSN 1471-4906. PMID 35090787.
- ↑ 10.00 10.01 10.02 10.03 10.04 10.05 10.06 10.07 10.08 10.09 10.10 10.11 10.12 10.13 10.14 10.15 10.16 10.17 10.18 10.19 10.20 10.21 10.22 10.23 10.24 Woodsworth, Daniel J.; Castellarin, Mauro; Holt, Robert A. (2013). "Sequence analysis of T-cell repertoires in health and disease". Genome Medicine 5 (10): 98. doi:10.1186/gm502. ISSN 1756-994X. PMID 24172704.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 11.7 11.8 Pai, Joy A.; Satpathy, Ansuman T. (August 2021). "High-throughput and single-cell T cell receptor sequencing technologies" (in en). Nature Methods 18 (8): 881–892. doi:10.1038/s41592-021-01201-8. ISSN 1548-7105. PMID 34282327.
- ↑ 12.0 12.1 Yeku, Oladapo; Frohman, Michael A. (2011). "Rapid Amplification of cDNA Ends (RACE)". RNA. Methods in Molecular Biology. 703. pp. 107–122. doi:10.1007/978-1-59745-248-9_8. ISBN 978-1-58829-913-0. https://pubmed.ncbi.nlm.nih.gov/21125486.
- ↑ Kuhn, Raphael; Sandu, Ioana; Agrafiotis, Andreas; Hong, Kai-Lin; Shlesinger, Danielle; Neimeier, Daniel; Merkler, Doron; Oxenius, Annette et al. (2022). "Clonally Expanded Virus-Specific CD8 T Cells Acquire Diverse Transcriptional Phenotypes During Acute, Chronic, and Latent Infections". Frontiers in Immunology 13: 782441. doi:10.3389/fimmu.2022.782441. ISSN 1664-3224. PMID 35185882.
- ↑ Waymack, James R.; Sundareshan, Vidya (2022), "Acquired Immune Deficiency Syndrome", StatPearls (Treasure Island (FL): StatPearls Publishing), PMID 30725978, http://www.ncbi.nlm.nih.gov/books/NBK537293/, retrieved 2023-02-28
- ↑ Pilkinton, Mark A.; McDonnell, Wyatt J.; Barnett, Louise; Chopra, Abha; Gangula, Rama; White, Katie D.; Leary, Shay; Currenti, Jennifer et al. (2021-03-25). "In Chronic Infection, HIV Gag-Specific CD4+ T Cell Receptor Diversity Is Higher than CD8+ T Cell Receptor Diversity and Is Associated with Less HIV Quasispecies Diversity". Journal of Virology 95 (8): e02380–20. doi:10.1128/JVI.02380-20. ISSN 0022-538X. PMID 33536169.
- ↑ Kervevan, Jérôme; Chakrabarti, Lisa A. (2021-01-07). "Role of CD4+ T Cells in the Control of Viral Infections: Recent Advances and Open Questions". International Journal of Molecular Sciences 22 (2): 523. doi:10.3390/ijms22020523. ISSN 1422-0067. PMID 33430234.
- ↑ 17.0 17.1 Hanahan, Douglas; Weinberg, Robert A. (2011-03-04). "Hallmarks of Cancer: The Next Generation" (in English). Cell 144 (5): 646–674. doi:10.1016/j.cell.2011.02.013. ISSN 0092-8674. PMID 21376230.
 |
