Biology:Test cross
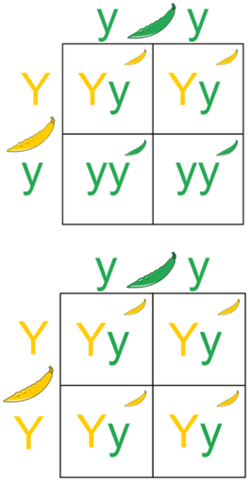
Under the law of dominance in genetics, an individual expressing a dominant phenotype could contain either two copies of the dominant allele (homozygous dominant) or one copy of each dominant and recessive allele (heterozygous dominant).[1] By performing a test cross, one can determine whether the individual is heterozygous or homozygous dominant.[1]
In a test cross, the individual in question is bred with another individual that is homozygous for the recessive trait and the offspring of the test cross are examined.[2] Since the homozygous recessive individual can only pass on recessive alleles, the allele the individual in question passes on determines the phenotype of the offspring.[3] Thus, this test yields 2 possible situations:
- If any of the offspring produced express the recessive trait, the individual in question is heterozygous for the dominant allele.[1]
- If all of the offspring produced express the dominant trait, the individual in question is homozygous for the dominant allele.[1]
History
The first uses of test crosses were in Gregor Mendel’s experiments in plant hybridization. While studying the inheritance of dominant and recessive traits in pea plants, he explains that the “signification” (now termed zygosity) of an individual for a dominant trait is determined by the expression patterns of the following generation.[4]
Rediscovery of Mendel’s work in the early 1900s led to an explosion of experiments employing the principles of test crosses. From 1908-1911, Thomas Hunt Morgan conducted test crosses while determining the inheritance pattern of a white eye-colour mutation in Drosophila.[5] These test cross experiments became hallmarks in the discovery of sex-linked traits.
Applications in model organisms
File:Caenorhabditis elegans 02.webm Test crosses have a variety of applications. Common animal organisms, called model organisms, where test crosses are often used include Caenorhabditis elegans and Drosophila melanogaster. Basic procedures for performing test crosses in these organisms are provided below:
C. elegans

To perform a test cross with C. elegans, place worms with a known recessive genotype with worms of an unknown genotype on an agar plate. Allow the male and hermaphrodite worms time to mate and produce offspring. Using a microscope, the ratio of recessive versus dominant phenotype will elucidate the genotype of the dominant parent.[6]
D. melanogaster
To perform a test cross with D. melanogaster, select a trait with a known dominant and recessive phenotype. Red eye colour is dominant and white is recessive. Obtain virgin females with white eyes, young males with red eyes, and put them into a single tube. Once offspring begin to appear as larvae, remove parental lines and observe the phenotype of adult offsprings.[7]
Limitations
There are many limitations to test crosses. It can be a time-consuming process as some organisms require a long growing time in each generation to show the necessary phenotype.[8] A large number of offspring are also required to have reliable data due to statistics.[9] Test crosses are only useful if dominance is complete. Incomplete dominance is when the dominant allele and recessive allele come together to form a blend of the two phenotypes in the offspring. Variable expressivity is when a single allele produces a range of phenotypes, which is also not accounted for in a test cross.
As more advanced techniques to determine genotype emerge, the test cross is becoming less prevalent in genetics. Genetic testing and genome mapping are modern advances which allow for more efficient and detailed information about one’s genotype to be determined.[10] Test crosses, however, are still used to this day and have created an excellent foundation for the development of more sophisticated techniques.
References
- ↑ 1.0 1.1 1.2 1.3 Gai, J.; He, J. (2013), Test Cross, Elsevier, pp. 49–50, doi:10.1016/b978-0-12-374984-0.01529-1, ISBN 978-0-08-096156-9, http://dx.doi.org/10.1016/b978-0-12-374984-0.01529-1, retrieved 2020-10-25
- ↑ Introduction to Genetic Analysis. New York: W.H. Freeman and Co.. 2005. pp. 34–40, 473–476, 626–629. ISBN 0-7167-4939-4. https://archive.org/details/introductiontoge00anth_210.
- ↑ Freeman, S; Harrington, M; Sharp, J (2014). "Using a Testcross to Confirm Predictions". Biological Science (Custom Edition for the University of British Columbia). Toronto, Ontario: Pearson Canada. pp. 260.
- ↑ Mendel, Gregor; Bateson, William (1925). Experiments in plant-hybridisation. Cambridge, Mass.: Harvard University Press. pp. 323–325. https://www.biodiversitylibrary.org/item/23469.
- ↑ "Thomas Hunt Morgan and the Discovery of Sex Linkage | Learn Science at Scitable" (in en). https://www.nature.com/scitable/topicpage/thomas-hunt-morgan-and-sex-linkage-452/.
- ↑ Fay, David S. (2018). "Classical genetic methods" (in en). WormBook: The Online Review of C. Elegans Biology (WormBook): 1–58. doi:10.1895/wormbook.1.165.1. PMID 24395816. PMC 4127492. https://www.ncbi.nlm.nih.gov/books/NBK179229/.
- ↑ Lawrence, Peter A. (1995). The making of a fly : the genetics of animal design. Oxford [England]: Blackwell Science. ISBN 0-632-03048-8. OCLC 24211238. https://www.worldcat.org/oclc/24211238.
- ↑ Orias, Eduardo (2012). "Chapter 10 - Tetrahymena thermophila Genetics: Concepts and Applications". Methods in Cell Biology. 109. Elsevier. pp. 301–325. doi:10.1016/B978-0-12-385967-9.00010-4. ISBN 978-0-12-385967-9.
- ↑ Lobo, I. "Genetics and Statistical Analysis | Learn Science at Scitable". https://www.nature.com/scitable/topicpage/genetics-and-statistical-analysis-34592/.
- ↑ Özgüç, Meral (2011). "Genetic testing: predictive value of genotyping for diagnosis and management of disease" (in en). EPMA Journal 2 (2): 173–179. doi:10.1007/s13167-011-0077-y. ISSN 1878-5077. PMID 23199147.
 |
