Biology:Transmembrane protein 255A
Transmembrane protein 255A[1] is a protein that is encoded by the TMEM255A gene.[2] TMEM255A is often referred to as family with sequence similarity 70, member A (FAM70A).[3] The TMEM255A protein is transmembrane and is predicted to be located the nuclear envelope of eukaryote organisms.[4]
Gene
The TMEM25A gene (often referred to as Family with Sequence Similarity 70 Member A; FAM70A) is located on Xq24, spanning 60,555 base pairs.[6] TMEM255A is flanked by the genes ATPase Na+/K+ transporting family member beta 4 (ATP1B4) and NFKB activating protein pseudogene 1 (NKAPP1).[7]
mRNA
There are three variants of the transcript seen, where isoform 1 is the longest. The 5’- and 3’- UTRs of the mRNA spans 227 and 2207 base pairs, respectively, and are predicted to contain several stem-loops.[8] The mRNA is 3512 base pairs long and the gene consists of 9 exons.[9]
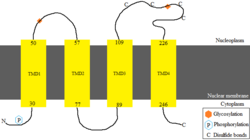
Protein
The longest protein encoded for is isoform 1, which spans 349 amino acids, and is predicted to have a molecular weight at 38 kDa and isoelectric point at pH 7.89.[11][12][13] Compared to the average vertebrate protein, TMEM255A is rich in aspartic acid, isoleucine, proline and tyrosine, and relatively poor in glutamic acid and lysine.[14] No charge clusters have been found in this protein.
The protein is predicted to be post-translationally modified by phosphorylation and glycosylation.[15] The protein is predicted to have four transmembrane domains in the nuclear membrane. The structure of the protein is predicted to be helical in the transmembrane domains.[16][17][18] Disulfide bonds are predicted to be found in the region in between transmembrane domains 3 and 4, which indicates that this particular region is located in the nucleoplasm.[19][20][21][22]
| Isoform | Accession number | Description | |
|---|---|---|---|
| 1 | NP_060408.3 | The longest transcript and isoform | |
| 2 | NP_001098014.1 | Shorter protein product than isoform 1, lacks one in-frame alternative midsection exon | |
| 3 | NP_001098015.1 | Lacks three in-frame exons. Shorter than isoform 1 and 2. |
Expression
TMEM255A is predicted to be most abundantly expressed in nerve, brain, testis, ovary, thymus and kidney. The protein is expressed in a variety of tissues, but at relatively moderate levels.[23][24][25]
Regulation of expression
Both the 5' and 3' Untranslated Regions (UTRs) are predicted to consist of several stem-loops.[26] The 3' UTR also contain a conserved miRNA target site (amino acids 22-29).[27] Phosphorylation and glycosylation sites have also been predicted in TMEM255A.[28][29]
Interacting proteins
Affinity Capture MS experimentally predicts that TMEM255A interacts with ten different proteins; Ankyrin repeat domain 13D (ANKRD13D), Collagen beta (1-O) galactosyltransferase 2 (COLGALT2), Grancalcin (GCA), Itchy E3 ubiquitin protein ligase (ITCH), Potassium channel tetramerization domain containing 2 (KCTD2), Neural precursor cell expressed developmentally down-regulated 4 (NEDD4), SEC24 family member B (SEC24D), Ubiquitin associated and SH3 domain containing B (UBASH3D), WW domain containing E3 ubiquitin protein ligase 1 and 2 (WWP1, WWP2) - most of these are included in ubiquitination processes, transcription regulation and protein degradation.[30]
Clinical significance
TMEM255A is predicted to be highly expressed in peroxisome proliferator-activated receptor γ coactivator 1α-upregulated glioblastoma multiforme cells (specific gene function not yet fully established).[31] Ongoing research is investigating the possibility of TMEM255A to be used in personalized immunotherapy.[32]
Homology
There is one known paralog for TMEM255A, called TMEM255B, which is found on chromosome 13 (position 13q34).[33] TMEM255A is only found in the kingdom of animalia, and its most distant homolog is found in invertebrata (i.e. Saccoglossus kowalenskii).
| Species | NCBI Accession # | Divergence (MYA) | Sequence Length (aa) | Sequence ID (%) | Sequence Similarity (%) |
|---|---|---|---|---|---|
| Homo sapiens (Human) | NP_060408.3 | - | 349 | 100 | 100 |
| Elephantulus edwardii (Cape Elephant Shrew) | XP_006893850.1 | 105 | 351 | 97 | 98 |
| Gallus gallus (Chicken) | XP_015134112.1 | 312 | 323 | 79 | 84 |
| Chrysemys picta bellii (Painted turtle) | XP_008167250.1 | 312 | 323 | 79 | 84 |
| Nanorana parkeri (High Himalaya frog) | XP_018415588.1 | 352 | 327 | 69 | 77 |
| Cyprinus carpio (Common carp) | XP_018971120.1 | 435 | 342 | 58 | 67 |
| Saccoglossus kowalevskii (Acorn worm) | XP_006819139.1 | 684 | 351 | 23 | 45 |
References
- ↑ Homo sapiens transmembrane protein 255A (TMEM255A), transcript variant - Nucleotide - NCBI. 2018-06-24. https://www.ncbi.nlm.nih.gov/nuccore/NM_017938.3.
- ↑ NCBI, National Center for Biotechnology Information. "TMEM255A Transmembrane Protein 255A [Homo sapiens"]. https://www.ncbi.nlm.nih.gov/gene/55026.
- ↑ Database, GeneCards Human Gene. "TMEM255A Gene - GeneCards | T255A Protein | T255A Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=TMEM255A.
- ↑ "Search: TMEM255A - The Human Protein Atlas". http://www.proteinatlas.org/search/TMEM255A.
- ↑ ] (2017-05-06). Image: GeneCards, 2017
- ↑ "TMEM255A". April 25, 2017. https://www.genecards.org/cgi-bin/carddisp.pl?gene=TMEM255A.
- ↑ "Gene neighbors for Gene (Select 55026) - Gene - NCBI". https://www.ncbi.nlm.nih.gov/gene?LinkName=gene_gene_neighbors&from_uid=55026.
- ↑ "The Mfold Web Server" (in EN). http://unafold.rna.albany.edu/?q=mfold.
- ↑ National Center for Biotechnology Information, NCBI. "TMEM255A transmembrane protein 255A [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/55026.
- ↑ Image: Kristin H. Aaen, 2017.
- ↑ National Center for Biotechnology Information, NCBI Gene (2017-04-02). "Transmembrane Domain 255A, Homo sapiens". https://www.ncbi.nlm.nih.gov/gene/55026.
- ↑ Subramaniam, S. (1998). "The Biology Workbench: a seamless database and analysis environment for the biologist". Proteins 2 (1): 1–2. doi:10.1002/(SICI)1097-0134(19980701)32:1<1::AID-PROT1>3.0.CO;2-Q. PMID 9672036.
- ↑ Toldo, Luca (April 25, 2017). "Gateway to Isoelectric Point Service". http://www.embl-heidelberg.de/cgi/pi-wrapper.pl.
- ↑ Dyer, K. F. (1971). "The quiet revolution: A new synthesis of biological knowledge". Journal of Biological Education 5: 15–24. doi:10.1080/00219266.1971.9653663. http://www.tiem.utk.edu/~gross/bioed/webmodules/aminoacid.htm. Retrieved 2017-05-06.
- ↑ Blom, N. (Summer 2002). "Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence". Proteomics 6: 1633–49.
- ↑ Yang, J. (2015). "The I-TASSER Suite: Protein structure and function prediction". Nature Methods 12 (1): 7–8. doi:10.1038/nmeth.3213. PMID 25549265.
- ↑ Roy, A. (2010). "I-TASSER: a unified platform for automated protein structure and function prediction". Nature Protocols 5 (4): 725–738. doi:10.1038/nprot.2010.5. PMID 20360767.
- ↑ Zhang, Y. (2008). "I-TASSER server for protein 3D structure prediction". BMC Bioinformatics 9: 40. doi:10.1186/1471-2105-9-40. PMID 18215316.
- ↑ Ferre & Clote (2006). "DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification". Nucleic Acids Research 34 (Web Server issue): W182-5. doi:10.1093/nar/gkl189. PMID 16844987.
- ↑ Ferre & Clote (Summer 2005). "DiANNA: a web server for disulfide connectivity prediction". Nucleic Acids Res. 33 (Web Server issue): W230–2. doi:10.1093/nar/gki412. PMID 15980459.
- ↑ Ferre & Clote (Summer 2005). "Disulfide connectivity prediction using secondary structure information and diresidue frequencies". Bioinformatics 21 (10): 2336–46. doi:10.1093/bioinformatics/bti328. PMID 15741247.
- ↑ Go (2010). "Redox control systems in the nucleus: mechanisms and functions". Antioxidants & Redox Signaling 13 (4): 489–509. doi:10.1089/ars.2009.3021. PMID 20210649.
- ↑ Input: TMEM255A. "EST Profile: Transmembrane Protein Domain 255A". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.437563.
- ↑ National Cancer Institute, Cancer Genome Anatomy Project. "Transmembrane protein 255A". https://cgap.nci.nih.gov/Genes/GeneInfo?ORG=Hs&CID=437563&LLNO=55026.
- ↑ BioGPS, BioGPS. "TMEM255A". http://biogps.org/#goto=genereport&id=55026.
- ↑ "SUNY Albany Research IT Group". 2017-04-02. http://unafold.rna.albany.edu/?q=mfold.
- ↑ Argawal (2017-04-17). "TargetScan Human: Prediction of miRNA Targets". http://www.targetscan.org/vert_71/.
- ↑ Gupta & Brunak (2002). "Prediction of glycosylation across the human proteome and the correlation to protein function". Pacific Symposium on Biocomputing 322: 310–22. PMID 11928486.
- ↑ Blom (2004-06-04). "Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence". Proteomics 4 (6): 1633–49. doi:10.1002/pmic.200300771. PMID 15174133.
- ↑ Chatr-Aryamontri (2016-12-14). "The BioGRID interaction database: 2017 update". Nucleic Acids Research 45 (D1): D369–D379. doi:10.1093/nar/gkw1102. PMID 27980099. PMC 5210573. https://thebiogrid.org/.
- ↑ Cho (2017-01-13). "Expression of PGC1α in glioblastoma multiforme patients". Oncology Letters 13 (6): 4055–76. doi:10.3892/ol.2017.5972. PMID 28599408.
- ↑ Weinschenk (2014). "Personalized immunotherapy against several neuronal and brain tumors". U.S. Patent Application No 14/531: 472. https://www.google.com/patents/CA2929445A1?cl=en&hl=no.
- ↑ Human Gene Database, GeneCards. "TMEM255B Gene". GeneCards. https://www.genecards.org/cgi-bin/carddisp.pl?gene=TMEM255B.
 |


