Biology:Triangle of U
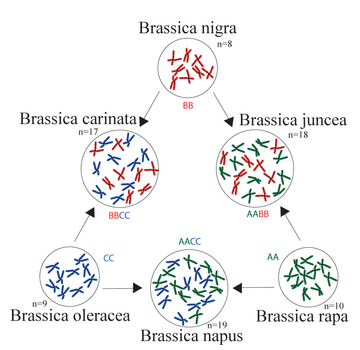
The triangle of U (/uː/ OO) is a theory about the evolution and relationships among the six most commonly known members of the plant genus Brassica. The theory states that the genomes of three ancestral diploid species of Brassica combined to create three common tetraploid vegetables and oilseed crop species.[1] It has since been confirmed by studies of DNA and proteins.[2]
The theory is summarized by a triangular diagram that shows the three ancestral genomes, denoted by AA, BB, and CC, at the corners of the triangle, and the three derived ones, denoted by AABB, AACC, and BBCC, along its sides.
The theory was first published in 1935 by Woo Jang-choon,[3] a Korean-Japanese botanist (writing under the Japanized name "U Nagaharu").[4] Woo made synthetic hybrids between the diploid and tetraploid species and examined how the chromosomes paired in the resulting triploids.
Woo's theory
The six species are
| Genomes | Chr. count | Species | Description |
|---|---|---|---|
| Diploid | |||
| AA | 2n=2x=20 | Brassica rapa | (syn. B. campestris) turnip, napa cabbage, bok choi |
| BB | 2n=2x=16 | Brassica nigra | black mustard |
| CC | 2n=2x=18 | Brassica oleracea | cabbage, kale, broccoli, Brussels sprouts, cauliflower, kohlrabi |
| Tetraploid | |||
| AABB | 2n=4x=36 | Brassica juncea | Brown mustard |
| AACC | 2n=4x=38 | Brassica napus | rapeseed, rutabaga |
| BBCC | 2n=4x=34 | Brassica carinata | Ethiopian mustard |
The code in the "Chr.count" column specifies the total number of chromosomes in each somatic cell, and how it relates to the number n of chromosomes in each full genome set (which is also the number found in the pollen or ovule), and the number x of chromosomes in each component genome. For example, each somatic cell of the tetraploid species Brassica napus, with letter tags AACC and count "2n=4x=38", contains two copies of the A genome, each with 10 chromosomes, and two copies of the C genome, each with 9 chromosomes, which is 38 chromosomes in total. That is two full genome sets (one A and one C), hence "2n=38" which means "n=19" (the number of chromosomes in each gamete). It is also four component genomes (two A and two C), hence "4x=38".[2]
The three diploid species exist in nature, but can easily interbreed because they are closely related. This interspecific breeding allowed for the creation of three new species of tetraploid Brassica.[3] (Critics, however, consider the geological separation too large.) These are said to be allotetraploid (containing four genomes from two or more different species); more specifically, amphidiploid (with two genomes each from two diploid species).[2]
Further relationships
The framework proposed by Woo, although backed by modern studies, leaves open questions about the time and place of hybridization and which species is the maternal or paternal parent. B. napus (AACC) is dated to have originated about 8,000[5] or 38,000–51,000[6] years ago. The homologous part of its constituent chromosomes has crossed over in many cultivars.[5] B. juncea (AABB) is estimated to have originated 39,000–55,000 years ago.[6] As of 2020, research on organellar genomes shows that B. nigra (BB) is likely the "mother" of B. carinata (BBCC) and that B. rapa (AA) likely mothered B. juncea. The situation with B. napus (AACC) is more complex: some specimens have a rapa-like organellar genome, while the rest indicate an ancient, unidentified maternal plant.[2]
Data from molecular studies indicate the three diploid species are themselves paleohexaploids.[7][8]
Allohexaploid species
In 2011 and 2018, novel allohexaploids (AABBCC) located at the "center" of the triangle of U were created by different means,[9][10][11] for example by crossing B. rapa (AA) with B. carinata (BBCC), or B. nigra (BB) with B. napus (AACC), or B. oleracea (CC) with B. juncea (AABB), followed by chromosome duplication of the triploid (ABC) offspring to generate doubled haploid (AABBCC) offspring.[11]
In addition, two stable allohexaploid (AABBSS) intergeneric hybrids between Indian mustard (B. juncea, AABB) and white mustard (Sinapis alba, SS) were created in 2020 by protoplast fusion.[12]
See also
References
- ↑ Jules, Janick (2009). Plant Breeding Reviews. 31. Wiley. p. 56. ISBN 978-0-470-38762-7. http://as.wiley.com/WileyCDA/WileyTitle/productCd-0470387629.html.
- ↑ 2.0 2.1 2.2 2.3 Xue, JY; Wang, Y; Chen, M; Dong, S; Shao, ZQ; Liu, Y (2020). "Maternal Inheritance of U's Triangle and Evolutionary Process of Brassica Mitochondrial Genomes.". Frontiers in Plant Science 11: 805. doi:10.3389/fpls.2020.00805. PMID 32595682. "Comparative genomic analyses can assign the subgenomes of the allotetraploids, B. juncea and B. napus, with their diploid parental taxa, and the results were in agreement with U’s triangle (Chalhoub et al., 2014; Yang et al., 2016a). [...]".
- ↑ 3.0 3.1 Nagaharu U (1935). "Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization". Japan. J. Bot 7: 389–452.
- ↑ "인터넷 과학신문 사이언스 타임즈" (in ko). http://junior.sciencetimes.co.kr/data/article/7000/0000006890.jsp.
- ↑ 5.0 5.1 Chalhoub, B; Denoeud, F; Liu, S; Parkin, IA; Tang, H; Wang, X; Chiquet, J; Belcram, H et al. (22 August 2014). "Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome.". Science 345 (6199): 950–3. doi:10.1126/science.1253435. PMID 25146293. https://www.researchgate.net/publication/267755510.
- ↑ 6.0 6.1 Yang, J; Liu, D; Wang, X; Ji, C; Cheng, F; Liu, B; Hu, Z; Chen, S et al. (October 2016). "The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection.". Nature Genetics 48 (10): 1225–32. doi:10.1038/ng.3657. PMID 27595476.
- ↑ Martin A. Lysak; Kwok Cheung; Michaela Kitschke; Petr Bu (October 2007). "Ancestral Chromosomal Blocks Are Triplicated in Brassiceae Species with Varying Chromosome Number and Genome Size" (PDF). Plant Physiology 145 (2): 402–10. doi:10.1104/pp.107.104380. PMID 17720758. PMC 2048728. http://www.plantphysiol.org/cgi/reprint/145/2/402. Retrieved 2010-08-22.
- ↑ Murat, Florent; Louis, Alexandra; Maumus, Florian; Armero, Alix; Cooke, Richard; Quesneville, Hadi; Crollius, Hugues Roest; Salse, Jerome (December 2015). "Understanding Brassicaceae evolution through ancestral genome reconstruction". Genome Biology 16 (1): 262. doi:10.1186/s13059-015-0814-y. PMID 26653025.
- ↑ Chen, Sheng; Nelson, Matthew N.; Chèvre, Anne-Marie; Jenczewski, Eric; Li, Zaiyun; Mason, Annaliese S.; Meng, Jinling; Plummer, Julie A. et al. (2011-11-01). "Trigenomic Bridges for Brassica Improvement". Critical Reviews in Plant Sciences 30 (6): 524–547. doi:10.1080/07352689.2011.615700. ISSN 0735-2689.
- ↑ Yang, Su; Chen, Sheng; Zhang, Kangni; Li, Lan; Yin, Yuling; Gill, Rafaqat A.; Yan, Guijun; Meng, Jinling et al. (2018-08-28). "A High-Density Genetic Map of an Allohexaploid Brassica Doubled Haploid Population Reveals Quantitative Trait Loci for Pollen Viability and Fertility". Frontiers in Plant Science 9: 1161. doi:10.3389/fpls.2018.01161. ISSN 1664-462X. PMID 30210508.
- ↑ 11.0 11.1 Gaebelein, Roman; Mason, Annaliese S. (2018-09-03). "Allohexaploids in the Genus Brassica". Critical Reviews in Plant Sciences 37 (5): 422–437. doi:10.1080/07352689.2018.1517143. ISSN 0735-2689.
- ↑ Kumari P, Singh KP, Kumar S, Yadava DK (2020). "Development of a Yellow-Seeded Stable Allohexaploid Brassica Through Inter-Generic Somatic Hybridization With a High Degree of Fertility and Resistance to Sclerotinia sclerotiorum.". Front Plant Sci 11: 575591. doi:10.3389/fpls.2020.575591. PMID 33329636.
 |
