Biology:ZNF548
 Generic protein structure example |
Zinc Finger Protein 548 (ZNF548) is a human protein encoded by the ZNF548 gene which is located on chromosome 19.[1] It is found in the nucleus and is hypothesized to play a role in the regulation of transcription by RNA Polymerase II. It belongs to the Krüppel C2H2-type zinc-finger protein family as it contains many zinc-finger repeats.[2]
Gene
This protein coding gene is 4987 bp long and encodes transcript variant 1 which is the longest ZNF548 isoform.[3][4] It is found on chromosome 19; its exact position is 19q13.43 in the plus strand. The gene has 4 exons which encode for a Kruppel associated box (KRAB domain) and 11 zinc finger repeats.[5][6]
Promoter
The promoter of the ZNF548 gene is 1198 bases long and is located at 57388850 - 57390047 on chromosome 19. The promoter region is conserved in 6 orthologs: Rhesus macaque (rhesus monkey), Pan troglodytes (chimpanzee), Oryctolagus cuniculus (European rabbit), Equus caballus (horse), Canis lupus familiaris (dog) and Sus scrofa (pig).[7]
Transcript

Transcript variant 1 is the longest transcript and encodes the longest protein isoform (ZNF548 isoform 1) which is 545 amino acids long.[3][4] Transcript variant 2 is missing exon 2 and encodes ZNF548 isoform 2 which is 533 amino acids long.[8][9]
Protein
ZNF548 belongs to the Kruppel C2H2-type zinc finger protein family as it contains 11 Cys2His2-type zinc finger repeats.[2] Each zinc finger has a conserved ββα structure where a zinc atom is fixed by C2H2 residues. ZNF548 is able to attach to the DNA at a 44 bp long sequence through its C2H2 Zn motifs, each binding to 4 DNA bases.[10] The protein also contains a Kruppel-associated box (KRAB) which is a domain found at the N terminus and contains multiple charged amino acids. This domain plays a role in transcription; it binds to the RING-B box-coiled coil (RBCC) domain of the KAP-1/TIF1-beta co-repressor.[11]
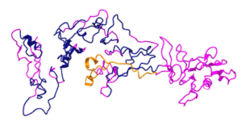
Tertiary structure of ZNF548 was predicted using I-Tasser.[13][14]
ZNF548 has a molecular weight of 64 kDa and a predicted isoelectric point of 8.21.[15] Compositional analysis of ZNF548 revealed that Alanine was found at a lower percentage than expected while Histidine was found at a much higher percentage than expected in the human ZNF548.[16]
Subcellular expression
The protein is found in the nucleus and could also be detected in the cytoplasm or the mitochondria.[17][18]
Tissue expression
ZNF548 expression in humans is relatively low compared to other proteins.[19] It has low tissue specificity; it is expressed and detected in all human tissues.[20]
Interactions with other proteins
Interaction of the human ZNF548 protein with the Nuclear distribution protein nudE-like 1 (NDEL1) and the Disrupted in schizophrenia 1 (DISC1) proteins has been experimentally validated using the two hybrid fragment pooling approach.[21] Leucine rich repeat containing 36 (LRRC36), Myotubularin 1 (MTM1), Myotubularin related protein 4 (MTMR4) and RNA binding motif protein 39 (RBM39) have also been detected to interact with the ZNF548 protein through Affinity Capture-Mass Spectrometry.[22]
Function
ZNF548 has been associated with gene expression as it can bind to nucleic acids as well as zinc ions. Based on the function of other KRAB-ZNF proteins it is hypothesized that it plays a role in the regulation of transcription of protein-encoding genes transcribed by RNA Polymerase II. Specifically it is a DNA-binding transcription factor that enhances or inhibits the transcription of certain genes that are transcribed by RNA Polymerase II. It has the ability to bind to a transcription factor recognition sequence that is on the same strand (cis) as the transcription start site via its zinc-fingers and become part of the KRAB-ZNF/KAP complex in the nucleoplasm. KRAB-ZNF proteins are known to be repressors. Therefore, when a KRAB-ZNF protein, such as ZNF548, is bound to DNA and simultaneously binds to the KAP1 co-repressor through its KRAB domain, various enzymes, such as histone deacetylases, histone methyltransferases and heterochromatin proteins, are recruited in order to compact the chromatin structure and consequently prevent transcription.[1][2][4][23][24][25]
Homologs
Orthologs
Orthologs of the ZNF548 protein have been found conserved across different orders of mammals only. This is line with the fact that C2H2-like fold groups are very common in mammalian transcription factors.[26]
The KRAB domain as well as the zinc finger repeats are highly conserved across orthologs.
| Genus, Species | Common name | Order | Estimated divergence date (MYA) | Accession number[27] | Sequence length (aa) | Sequence similarity to human protein (%) |
|---|---|---|---|---|---|---|
| Homo sapiens | Human | Primates | 0 | NP_001166244.1 | 545 | 100 |
| Pan troglodytes | Chimpanzee | Primates | 6.7 | XP_003316775.1 | 545 | 99.3 |
| Rhinopithecus roxellana | Golden snub-nosed monkey | Primates | 29.44 | XP_010385311.1 | 545 | 97.8 |
| Cercocebus atys | Sooty mangabey | Primates | 29.44 | XP_011931548.1 | 545 | 97.6 |
| Trachypithecus francoisi | François' langur | Primates | 29.44 | XP_033080506.1 | 545 | 97.8 |
| Oryctolagus cuniculus | European rabbit | Lagomorpha | 90 | XP_017193400.1 | 532 | 84.2 |
| Marmota monax | Groundhog | Rodentia | 90 | KAF7471703.1 | 546 | 84.9 |
| Ictidomys tridecemlineatus | Thirteen-lined ground squirrel | Rodentia | 90 | XP_013221367.2 | 546 | 84.9 |
| Octodon degus | Common degu | Rodentia | 90 | XP_012368689.1 | 548 | 81.1 |
| Castor canadensis | North American beaver | Rodentia | 90 | XP_020025721.1 | 545 | 76 |
| Heterocephalus glaber | Naked mole-rat | Rodentia | 90 | XP_012921729.1 | 599 | 77.1 |
| Enhydra lutris kenyoni | Sea otter | Carnivora | 96 | XP_022347780.1 | 583 | 76 |
| Leptonychotes weddellii | Weddell seal | Carnivora | 96 | XP_030883405.1 | 634 | 66.4 |
| Ailuropoda melanoleuca | Giant panda | Carnivora | 96 | XP_034495390.1 | 630 | 65.6 |
| Equus przewalskii | Przewalski's horse | Perissodactyla | 96 | XP_008522443.1 | 579 | 71.4 |
| Ceratotherium simum simum | Southern white rhinoceros | Perissodactyla | 96 | XP_014649872.1 | 578 | 71.1 |
| Physeter catodon | Sperm whale | Artiodactyla | 96 | XP_023972760.1 | 589 | 72.5 |
| Balaenoptera musculus | Blue whale | Artiodactyla | 96 | XP_036688369.1 | 592 | 72.3 |
| Lipotes vexillifer | Baiji | Artiodactyla | 96 | XP_007457531.1 | 582 | 72.1 |
| Bos taurus | Cattle | Artiodactyla | 96 | NP_001193737.1 | 581 | 70.3 |
| Pteropus alecto | Black flying fox | Chiroptera | 96 | XP_024905354.1 | 678 | 64.1 |
Paralogs
ZNF548 has 25 paralogous proteins in human as seen in the table below.
| Protein name | Accession number | Sequence similarity to human ZNF548 protein (%) |
|---|---|---|
| ZNF548 | NP_001166244.1 | 100 |
| ZIK1 | NP_001010879.2 | 57.6 |
| ZNF792 | NP_787068.3 | 56.2 |
| ZNF419 | NP_001091961.1 | 56.2 |
| ZNF154 | NP_001078853.1 | 55.3 |
| ZNF256 | NP_005764.2 | 54.7 |
| ZNF549 | NP_001186224.2 | 52.9 |
| ZNF586 | NP_060122.2 | 52.7 |
| ZNF773 | NP_940944.1 | 52.2 |
| ZNF418 | NP_001303956.1 | 51.6 |
| ZNF480 | NP_653285.2 | 51.6 |
| ZNF551 | NP_612356.2 | 50.7 |
| ZNF304 | NP_001277247.1 | 50.3 |
| ZNF583 | NP_001153332.1 | 49.2 |
| ZNF570 | NP_001287922.1 | 48.9 |
| ZNF772 | NP_001019767.1 | 48.6 |
| ZNF587B | NP_001363152.1 | 48.6 |
| ZNF79 | NP_009066.2 | 48.4 |
| ZNF8 | NP_066575.2 | 46.4 |
| ZNF584 | NP_775819.1 | 46 |
| ZNF561 | NP_689502.2 | 45.8 |
| ZNF552 | NP_079038.2 | 43 |
| ZNF610 | NP_001154897.1 | 42.6 |
| ZNF793 | NP_001013681.2 | 41.7 |
| ZNF562 | NP_001123503.1 | 41 |
| ZNF691 | NP_001229668.1 | 31.5 |
Clinical Significance
ZNF548 was identified as a gene in meta-virus signature (MVS) which can be used to distinguish individuals with viral infections from those with bacterial infections as well as from healthy individuals.[28]
ZNF548 microRNA expression can act as a marker to diagnose ovarian cancer.[29] ZNF548 blood gene expression biomarker can also be used as a marker for suicidality.[30]
References
- ↑ 1.0 1.1 "ZNF548 Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=ZNF548.
- ↑ 2.0 2.1 2.2 "UniProtKB - Q8NEK5 (ZN548_HUMAN)". https://www.uniprot.org/uniprot/Q8NEK5#function.
- ↑ 3.0 3.1 Homo sapiens zinc finger protein 548 (ZNF548), transcript variant 1, mRNA. 11 December 2020. https://www.ncbi.nlm.nih.gov/nuccore/NM_001172773.2.
- ↑ 4.0 4.1 4.2 "zinc finger protein 548 isoform 1 [Homo sapiens - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein/289666756.
- ↑ "Transcript: ZNF548-201 (ENST00000336128.12) - Protein summary - Homo_sapiens - Ensembl genome browser 104". http://useast.ensembl.org/Homo_sapiens/Transcript/ProteinSummary?g=ENSG00000188785;r=19:57389854-57402992;t=ENST00000336128.
- ↑ "ZNF548 Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=ZNF548.
- ↑ "Promoter region for human ZNF548 gene". https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=d51bcdb36fcf9ebaf336786862f78ca0;SHOW_ANNOTATION=ZNF17---TRAPPC2B---Z;ELDORADO_VERSION=E36R2011.
- ↑ Homo sapiens zinc finger protein 548 (ZNF548), transcript variant 2, mRNA. 18 December 2020. https://www.ncbi.nlm.nih.gov/nuccore/NM_152909.4.
- ↑ "zinc finger protein 548 isoform 2 [Homo sapiens"]. https://www.ncbi.nlm.nih.gov/protein/NP_690873.2.
- ↑ "Zinc finger C2H2 superfamily". http://www.ebi.ac.uk/interpro/entry/InterPro/IPR036236/.
- ↑ "Kruppel-associated box". http://www.ebi.ac.uk/interpro/entry/InterPro/IPR001909/.
- ↑ "I-TASSER server for protein structure and function prediction". https://zhanggroup.org/I-TASSER/.
- ↑ J Yang, Y Zhang. I-TASSER server: new development for protein structure and function predictions, Nucleic Acids Research, 43: W174-W181, 2015.
- ↑ C Zhang, PL Freddolino, Y Zhang. COFACTOR: improved protein function prediction by combining structure, sequence and protein–protein interaction information. Nucleic Acids Research, 45: W291-W299, 2017.
- ↑ "Compute pI/MW of ZNF548". https://www.expasy.org/resources/compute-pi-mw.
- ↑ "Statistical Analysis of Protein Sequence for ZNF548". https://www.ebi.ac.uk/Tools/seqstats/saps/.
- ↑ "Prediction of eukaryotic protein subcellular localization using deep learning for ZNF548". https://services.healthtech.dtu.dk/service.php?DeepLoc-1.0.
- ↑ "PSORT II Prediction for human ZNF548 protein". https://psort.hgc.jp/form2.html.
- ↑ "Protein Abundance Database entry on ZNF548". https://pax-db.org/protein/1852017.
- ↑ "The Human Protein Atlas entry on ZNF548". https://www.proteinatlas.org/ENSG00000188785-ZNF548.
- ↑ Sügis E, Dauvillier J, Leontjeva A, et al. HENA, heterogeneous network-based data set for Alzheimer's disease. Scientific Data. 2019 Aug;6(1):151. doi:10.1038/s41597-019-0152-0. PMID: 31413325; PMCID: PMC6694132.
- ↑ "ZNF548 interactions with other proteins". https://thebiogrid.org/127076/summary/homo-sapiens/znf548.html.
- ↑ Gaudet, P., Livstone, M. S., Lewis, S. E., & Thomas, P. D. (2011). Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Briefings in Bioinformatics, 12(5), 449–462. doi:10.1093/bib/bbr042
- ↑ Reactome entry on Transcription regulation [https://reactome.org/PathwayBrowser/#/R-HSA-212436&FLG=Q8NEK5]
- ↑ Reactome entry on KRAB-ZNF/KAP interaction [https://reactome.org/content/detail/R-HSA-975040]
- ↑ "Zinc fingers C2H2-type". https://www.genenames.org/data/genegroup/#!/group/28.
- ↑ "BLAST of ZNF548 protein". https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ Andres-Terre, M. (2018). Exploring the Heterogeneity of Immune Response to Viral and Bacterial Infection. ProQuest Dissertations Publishing.
- ↑ Aboutalebi, H., et al. (2020). The diagnostic, prognostic and therapeutic potential of circulating microRNAs in ovarian cancer. The International Journal of Biochemistry & Cell Biology, 124, 105765. doi:10.1016/j.biocel.2020.105765
- ↑ Levey, D. et al. (2016). Towards understanding and predicting suicidality in women: Biomarkers and clinical risk assessment. Molecular Psychiatry, 21(6), 768–785. doi:10.1038/mp.2016.31
 |
