Chemistry:Chlorotonil A
| |
| Names | |
|---|---|
| IUPAC name
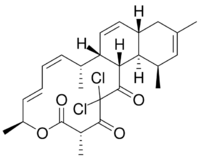
(1S,2R,3Z,5E,7S,10S,14R,15R,16S,20S)-12,12-dichloro-2,7,10,16,18-pentamethyl-8-oxatricyclo[12.8.0.015,20]docosa-3,5,17,21-tetraene-9,11,13-trione
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEMBL | |
| ChemSpider | |
PubChem CID
|
|
| |
| |
| Properties | |
| C26H32Cl2O4 | |
| Molar mass | 479.436 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Chlorotonil A is a polyketide natural product produced by the myxobacterium Sorangium cellulosum So ce1525.[2] It displays antimalarial activity in an animal model,[3] and has in vitro antibacterial and antifungal activity.[citation needed] The activity of chlorotonil A has been attributed to the gem-dichloro-1,3-dione moiety, which is a unique functionality in polyketides. In addition to its unique halogenation, the structure of chlorotonil A has also garnered interest due to its similarity to anthracimycin, a polyketide natural product with antibiotic activity against Gram-positive bacteria.
Biosynthesis
Chlorotonil A is synthesized from a type I modular polyketide synthase (PKS). This gene cluster does not have any acyltransferase (AT) domains, indicating that it is a trans-AT PKS; in these systems, there is a tandem-AT domain that loads the extender subunits onto the acyl carrier protein (ACP) and checks the intermediates, rather than individual AT domains in each module. The gene cluster of chlorotonil A is organized so that the initiator, acetyl-CoA, is loaded onto the tandem-AT domain, then is iteratively elongated with malonyl-CoA units to construct the macrolactone backbone. At modules 3 and 7, a double bond shift occurs in the elongation module to allow for the β,γ-unsaturation and α-methylation. There is a spontaneous, non-enzymatic intramolecular Diels-Alder-like [4+2] cycloaddition at module 8 to furnish the decalin motif.
Following macrolactonization by the thioesterase domain of module 10, the premature chlorotonil A core is chlorinated twice by CtoA, a flavin-dependent halogenase. The halogenated core is then methylated by the standalone SAM-dependent methyltransferase CtoF to yield chlorotonil A.[4]
References
- ↑ "Chlorotonil A | Chemical Substance Information | J-GLOBAL" (in en). https://jglobal.jst.go.jp/en/detail?JGLOBAL_ID=200907027847718540.
- ↑ Gerth, Klaus; Steinmetz, Heinrich; Höfle, Gerhard; Jansen, Rolf (2008). "Chlorotonil A, a Macrolide with a Unique gem-Dichloro-1,3-dione Functionality from Sorangium cellulosum, So ce1525". Angewandte Chemie International Edition 47 (3): 600–602. doi:10.1002/anie.200703993. PMID 18058875.
- ↑ Held, Jana; Gebru, Tamirat; Kalesse, Markus; Jansen, Rolf; Gerth, Klaus; Müller, Rolf; Mordmüller, Benjamin (2014). "Antimalarial Activity of the Myxobacterial Macrolide Chlorotonil A". Antimicrobial Agents and Chemotherapy 58 (11): 6378–6384. doi:10.1128/AAC.03326-14. PMID 25114138.
- ↑ Jungmann, Katrin; Jansen, Rolf; Gerth, Klaus; Huch, Volker; Krug, Daniel; Fenical, William; Müller, Rolf (2015). "Two of a Kind— The Biosynthetic Pathways of Chlorotonil and Anthracimycin". ACS Chemical Biology 10 (11): 2480–2490. doi:10.1021/acschembio.5b00523. PMID 26348978.
See also
 |



