Chemistry:Disorazol

| |
| Names | |
|---|---|
| IUPAC name
(2Z,4E,9Z,23Z,25Z)-12,28-bis[(E)-3-hydroxy-2-methylhex-4-en-2-yl]-20-methoxy-7,13,17,29,33-pentaoxa-34,35-diazatetracyclo[29.2.1.115,18.06,8]pentatriaconta-1(34),2,4,9,15,18(35),21,23,25,31-decaene-14,30-dione
| |
| Other names
Disorazole; Disorazol A
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider |
|
PubChem CID
|
|
| |
| |
| Properties | |
| C43H54N2O10 | |
| Molar mass | 758.909 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Disorazol, a cyclic polyketide synthesized by the bacterium Sorangium cellulosum So ce12, was first detected and isolated in 1994.[1] Its chemical structure consists of a macrocyclic ring and two oxazole rings.[1] Disorazol A has been demonstrated to exhibit anti-fungi activities, but it was not active against yeasts.[1] In addition, this substance demonstrates potent anti-cancer characteristics at exceptionally low picomolar levels by obstructing the mechanism of tubulin assembly and triggering the disruption of microtubules. As a result, these impacts lead to the initiation of cell apoptosis.[2] However, disorazols cannot be directly used as drugs in the clinic due to its extremely high cytotoxicity and instability.[2] Thus, chemical and biosynthetic synthesis pathways were designed to synthesize unnatural derivatives of disorazol in hope of reducing its cytotoxicity without decreasing its anti-cancer potency.[2]
Biosynthesis
Four synthetic genes, disABCD, have been identified and documented as contributors to the biosynthesis of disorazol. The disABC genes encode hybrid trans-AT type polyketide synthase (PKS) megaenzymes, while the disD gene encodes an additional acyl transferase protein and an enoylreductase (ER).[2] Notably, the ER encoded in the disD gene was not involved in the biosynthesis pathway of disorazol.[2]
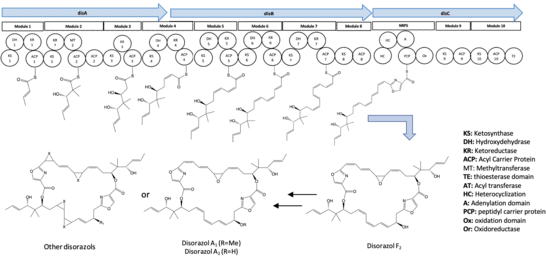
Generally, 10 synthetic modules were involved in the biosynthesis pathway of disorazol. The formation of half of the disorazol bis-lactone core involves the utilization of seven malonyl-CoA molecules and one serine molecule as extender units.[3] Subsequently, the dimerization or cyclization of two polyketide monomers takes place within the thioesterase (TE) domain located in module 10, leading to the synthesis of disorazol.[3] Furthermore, the release of the final product is facilitated by module 8, 9, and 10.[3] The process of PKS monomer dimerization in the biosynthesis pathway of disorazol exhibits an unconventional nature.[2]
References
- ↑ 1.0 1.1 1.2 Irschik, Herbert; Jansen, Rolf; Gerth, Klaus; HöFle, Gerhard; Reichenbach, Hans (1995). "Antibiotics from gliding bacteria. No.62. Disorazol A, an Efflcient Inhibitor of Eukaryotic Organisms Isolated from Myxobacteria." (in en). The Journal of Antibiotics 48 (1): 31–35. doi:10.7164/antibiotics.48.31. ISSN 0021-8820. PMID 7868386. http://joi.jlc.jst.go.jp/JST.Journalarchive/antibiotics1968/48.31?from=CrossRef.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Wang, Zong-Jie; Liu, Xiaotong; Zhou, Haibo; Liu, Yang; Tu, Qiang; Huo, Liujie; Yan, Fu; Müller, Rolf et al. (2023-04-21). "Engineered Biosynthesis of Complex Disorazol Polyketides in a Streamlined Burkholderia thailandensis" (in en). ACS Synthetic Biology 12 (4): 971–977. doi:10.1021/acssynbio.2c00610. ISSN 2161-5063. PMID 36988632. https://pubs.acs.org/doi/10.1021/acssynbio.2c00610.
- ↑ 3.0 3.1 3.2 Tu, Qiang; Herrmann, Jennifer; Hu, Shengbiao; Raju, Ritesh; Bian, Xiaoying; Zhang, Youming; Müller, Rolf (2016-02-15). "Genetic engineering and heterologous expression of the disorazol biosynthetic gene cluster via Red/ET recombineering" (in en). Scientific Reports 6 (1): 21066. doi:10.1038/srep21066. ISSN 2045-2322. PMID 26875499.
 |

