Chemistry:Lactocillin
| |
| Names | |
|---|---|
| IUPAC name
N-({2'-[(26Z)-26-Ethyliden-19,29-bis(1-hydroxyethyl)-12-{[(1H-indol-3-ylcarbonyl)sulfanyl]methyl}-14,21,28,31-tetraoxo-10,17,24,34-tetrathia-6,13,20,27,30,35,36,37,38-nonaazahexacyclo[30.2.1.18,11.1
15,18.122,25.02,7]octatriaconta-1(35),2,4,6,8,11(38),18(37),22,25(36),32-decaen-5-yl]-2,4'-bi-1,3-thiazol-4-yl}carbonyl)alanin
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider | |
PubChem CID
|
|
| |
| |
| Properties | |
| C51H45N13O10S7 | |
| Molar mass | 1224.42 g·mol−1 |
| Density | 1.8 g/cm3 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Lactocillin is a thiopeptide antibiotic which is encoded for and produced by biosynthetic genes clusters in the bacteria Lactobacillus gasseri. Lactocillin was discovered and purified in 2014.[1] Lactobacillus gasseri is one of the four Lactobacillus bacteria found to be most common in the human vaginal microbiome.[2] Due to increasing levels of pathogenic resistance to known antibiotics, novel antibiotics are increasingly valuable. Lactocillin could function as a new antibiotic that could help people fight off infections that are resistant to many other antibiotics.
Biosynthetic Gene Clusters
Lactocillin is produced by a biosynthetic gene cluster, which is a group of genes in bacteria that work together to make a secondary metabolite.[3] Secondary metabolites are molecules with many different chemical structures and functions, and in this case, lactocillin functions as an antibiotic.[3]
Biosynthetic gene clusters are similar to operons in bacteria in that they both code for proteins that function together in a common process. However, biosynthetic gene clusters always code for a known secondary metabolite, while operons are a general group of genes under one promoter. Operons can code for a specific molecule, similar to BGC’s, or other things such as associated proteins that work together in a common function, such as lac operon coding for the proteins involved in breaking down lactose.[citation needed]
Lactocillin is made by biosynthetic gene cluster 66 (bgc66) which is located on a plasmid in Lactobacillus gasseri.[1] bgc66 has many different genes that code for the proteins shown in the table below and perform the indicated function involved in the synthesis of lactocillin.[1]
| Protein | Function |
| YcaO | Aids in heterocycle formation, as we see five cysteine derived heterocycles in lactocillin.[4] |
| cyclodehydratase | Aids in heterocycle formation, as we see five cysteine derived heterocycles in lactocillin.[4] |
| Lantibiotic DH (2 different genes of this type) | Makes dehydrobutyrine residues, as one is seen in lactocillin.[5] |
| TclM | Aids in the production of the trithiazoylpyridine core of lactocillin.[1] |
| Other post-translation modification enzymes | Help continue to adjust the structure of lactocillin after translation. |
| Regulatory proteins | Regulate transcription of the BGC.[1] |
Thiopeptides
Antibiotics are chemicals used to inhibit or kill microbes, and come in many different chemical classes. Thiopeptides are a fairly new chemical class of antibiotics, characterized by a central six-membered ring with a nitrogen in the ring. Certain thiopeptides are created by bacteria found in other unusual places such as marine life and soil, but lactocillin is made by bacteria found in human vaginal and oral microbiomes.[1]
Thiopeptides work well against gram positive bacteria but not gram negative bacteria.[1] Thiopeptides have even been found to be potentially effective in fighting MRSA.[6] Thiopeptides are seen to potentially have many functions such as “anticancer, antiplasmodial, immunosuppressive, renin inhibitory, RNA polymerase inhibitory, and antifungal activities”.[7]
Thiopeptides function as antibiotic by blocking ribosomal protein synthesis.[7] This is an example of post-transcriptional regulation, as the thiopeptides do not affect transcription of the proteins but do prevent translation.[citation needed]
Lactocillin is different from thiocillin (a well-studied thiopeptide) in 3 important ways. Lactocillin 1) has a free carboxylic acid at C-terminus, 2) doesn’t undergo any post-translation modifications that require oxygen, and 3) has an indolyl-S-cysteine residue at position 8.[1] These differences suggest that this thiopeptide may function differently than the others, but more research needs to be done to determine if this is accurate.
Structure
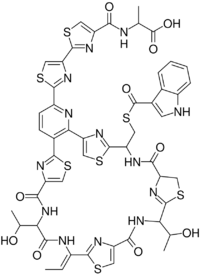
Lactocillin has an empirical formula of C51H45N13O10S7 and has a picture as shown above, as determined using NMR and UV-Vis absorbance profiles.[1] The structure of this protein does not perfectly match up to the sequence of bcg66. This illustrates that there must be some sort of post-translational modification.[1]
Plasmid harboring and Horizontal Gene Transfer
Horizontal gene transfer (HGT) is the mechanism by which bacteria can share genes, besides from parent to offspring in reproduction. Bacteria can add genes to their genome that may improve their fitness by taking in genetic material from other bacteria, the environment, or bacteriophages through HGT.[citation needed]
bgc66 is located on a plasmid along with other maintenance, regulatory, transfer, and transposases sequences.[1] The presence of these transfer sequences shows us that this plasmid can participate in HGT leading to other bacteria being able to produce lactocillin. The transposase sequences suggest that the plasmid might have even further capabilities of gene transfer through the cutting out and insertion of certain genes through the use of transposase proteins. If lactocillin is further studied and determined to have desirable properties, the fact that the BGC for lactocillin’s production resides on a transferable plasmid would make it easier for chemical companies to mass produce it.[citation needed]
Function in Human Body
Lactocillin is seen to be very functional in killing certain pathogens.[1] The minimum inhibitory concentration (MIC) of an antibiotic is the lowest concentration of an antibiotic needed to inhibit the growth of a bacteria. The MIC was calculated for many different bacteria that commonly infect human vaginas. This approach makes sense, as a non-pathogenic bacterium found in the vaginal microbiota might provide a benefit to the host, such as resistance to potential pathogens that commonly infect that area of the body, because the host’s survival is essential for the bacteria’s survival.[citation needed]
Growth was observed at multiple different concentrations to obtain the MIC of lactocillin for different pathogens. Lactocillin was found to be most effective at preventing growth of Staphylococcus aureus, Corynebacterium aurimucosum, and Streptococcus sobrinus, but was also effective at inhibiting growth at higher concentrations for other bacterial pathogens. These well inhibited bacteria cause Staph infections, urinary tract infections,[8] and cavities.[9] This suggests that lactocillin could potentially be used as a common antimicrobial in the future.[citation needed]
Lactocillin was not seen to prevent growth of other bacteria that are known to be common and beneficial to the vaginal microbiome. This makes sense, as killing these bacteria would be deleterious to a human’s health and, in consequence, also deleterious to the bacteria’s chance at survival.[10]
Other Lactobacillus bacteria have been seen to also be used as probiotics.[11] Lactobacillus gasseri could potentially function similarly to these other Lactobacillus bacteria and be used as a probiotic, helping with overall health such as immunity, cholesterol levels, and skin health.[11]
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Donia, Mohamed S.; Cimermancic, Peter; Schulze, Christopher J.; Wieland Brown, Laura C.; Martin, John; Mitreva, Makedonka; Clardy, Jon; Linington, Roger G. et al. (2014-09-11). "A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics". Cell 158 (6): 1402–1414. doi:10.1016/j.cell.2014.08.032. ISSN 1097-4172. PMID 25215495.
- ↑ Vásquez, Alejandra; Jakobsson, Tell; Ahrné, Siv; Forsum, Urban; Molin, Göran (August 2002). "Vaginal lactobacillus flora of healthy Swedish women". Journal of Clinical Microbiology 40 (8): 2746–2749. doi:10.1128/jcm.40.8.2746-2749.2002. ISSN 0095-1137. PMID 12149323.
- ↑ 3.0 3.1 Chen, Ray; Wong, Hon Lun; Burns, Brendan Paul (2019-02-25). "New Approaches to Detect Biosynthetic Gene Clusters in the Environment". Medicines 6 (1): 32. doi:10.3390/medicines6010032. ISSN 2305-6320. PMID 30823559.
- ↑ 4.0 4.1 Dunbar, Kyle L.; Melby, Joel O.; Mitchell, Douglas A. (2012-04-22). "YcaO domains use ATP to activate amide backbones during peptide cyclodehydrations". Nature Chemical Biology 8 (6): 569–575. doi:10.1038/nchembio.944. ISSN 1552-4469. PMID 22522320.
- ↑ Ortega, Manuel A.; Hao, Yue; Zhang, Qi; Walker, Mark C.; van der Donk, Wilfred A.; Nair, Satish K. (2015-01-22). "Structure and Mechanism of the tRNA-Dependent Lantibiotic Dehydratase NisB". Nature 517 (7535): 509–512. doi:10.1038/nature13888. ISSN 0028-0836. PMID 25363770. Bibcode: 2015Natur.517..509O.
- ↑ Haste, Nina M.; Thienphrapa, Wdee; Tran, Dan N.; Loesgen, Sandra; Sun, Peng; Nam, Sang-Jip; Jensen, Paul R.; Fenical, William et al. (December 2012). "Activity of the thiopeptide antibiotic nosiheptide against contemporary strains of methicillin-resistant Staphylococcus aureus". The Journal of Antibiotics 65 (12): 593–598. doi:10.1038/ja.2012.77. ISSN 1881-1469. PMID 23047246.
- ↑ 7.0 7.1 Just-Baringo, Xavier; Albericio, Fernando; Álvarez, Mercedes (2014-01-17). "Thiopeptide antibiotics: retrospective and recent advances". Marine Drugs 12 (1): 317–351. doi:10.3390/md12010317. ISSN 1660-3397. PMID 24445304.
- ↑ Lo, Seynabou; Thiam, Issa; Fall, Bécaye; Ba-Diallo, Awa; Diallo, Oumarou Foly; Diagne, Rokhaya; Dia, Mamadou Lamine; Ka, Roughyatou et al. (2015-07-14). "Urinary tract infection with Corynebacterium aurimucosum after urethroplasty stricture of the urethra: a case report". Journal of Medical Case Reports 9: 156. doi:10.1186/s13256-015-0638-0. ISSN 1752-1947. PMID 26155836.
- ↑ Conrads, Georg; de Soet, Johannes J.; Song, Lifu; Henne, Karsten; Sztajer, Helena; Wagner-Döbler, Irene; Zeng, An-Ping (2014). "Comparing the cariogenic species Streptococcus sobrinus and S. mutans on whole genome level". Journal of Oral Microbiology 6: 26189. doi:10.3402/jom.v6.26189. ISSN 2000-2297. PMID 25475081.
- ↑ Milshteyn, Aleksandr; Colosimo, Dominic A.; Brady, Sean F. (13 June 2018). "Accessing Bioactive Natural Products from the Human Microbiome". Cell Host & Microbe 23 (6): 725–736. doi:10.1016/j.chom.2018.05.013. ISSN 1934-6069. PMID 29902438.
- ↑ 11.0 11.1 Reid, Gregor (1999-09-01). "The Scientific Basis for Probiotic Strains ofLactobacillus" (in en). Applied and Environmental Microbiology 65 (9): 3763–3766. doi:10.1128/AEM.65.9.3763-3766.1999. ISSN 1098-5336. PMID 10473372. Bibcode: 1999ApEnM..65.3763R.
 |
