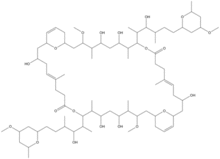
Chemistry:Swinholide
| |
| Identifiers | |
|---|---|
| |
3D model (JSmol)
|
|
| ChEMBL |
|
| ChemSpider | |
PubChem CID
|
|
| |
| |
| Properties | |
| C78H132O20 | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Swinholides are dimeric 42 carbon-ring polyketides that exhibit a 2-fold axis of symmetry. Found mostly in the marine sponge Theonella, swinholides encompass cytotoxic and antifungal activities via disruption of the actin skeleton.[1] Swinholides were first described in 1985[2] and the structure and stereochemistry were updated in 1989[3] and 1990,[4] respectively. Thirteen swinholides have been described in the literature, including close structural compounds such as misakinolides/bistheonellides,[5] ankaraholides,[6] and hurgholide A[7] It is suspected that symbiotic microbes that inhabit the sponges rather than the sponges themselves produce swinholides since the highest concentration of swinholides are found in the unicellular bacterial fraction of sponges and not in the sponge fraction or cyanobacteria fraction that also inhabit the sponges.[4][8][9]
From a marine field sample containing the cyanobacterium Symploca sp, Swinholide A has also been reported in literature.[6] The structural analogs of swinholides, ankaraholides, were also found from the cyanobacterium Geitlerinema sp. in the same experimental study.[6] Since sponges host a range of bacteria, including symbiotic cyanobacteria, it is often wondered how swinholides are produced.[1] A study of the production of misakinolide revealed that it was attributed to the Theonella symbiont bacterium Candidatus Entotheonella via the discovery of a trans-AT polyketides synthase (PKS) biosynthesis gene cluster.[1] This demonstrates that the true origin of swinholides is symbiotic bacteria that inhabit sponges.[1]
History
Cyanobacteria are known to have a wide application scope due to their structurally varied secondary metabolites they produce.[1] Among many of the secondary metabolites, polyketides have demonstrated vital bioactivities that can be applied to a number of fields. For example, many antifungal, antitumor, and antibiotic polyketides are found from plants, bacteria, and fungi.[1] The synthesis of polyketides is well known: small monomeric compounds frame polyketides via elongation on multidomain PKS complexes.[1] The PKSs can add either a malonyl, acyl, or derivative unit to the chain and they are classified into types 1-3, which are dependent on factors such as functionality and domain architecture.[1] Type I PKSs include cis- and trans-acyltransferase (trans-AT) PKSs where each section of the cis-AT PKSs encodes a dedicated AT domain and the trans-AT PKSs have distinct ATs that are used in place of the cis-encoded AT domains.[1]
Structure
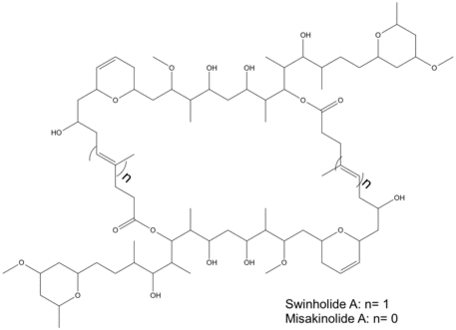
The structures of swinholide, misakinolide, and luminaolide are shown below. (Figure 1 and Figure 2).[1]

Biosynthesis
The swinholide biosynthesis gene cluster (swi) was located on a single scaffold by BLASTp searches against misakinolide biosynthesis cluster genes. This was chosen because of the close structural resemblance of these compounds.[10]
The swinholide biosynthesis gene cluster (85-kb) encodes for five PKS proteins, including SwiC to SwiG.[1] This includes an AT enzyme, SwiG, which is an characteristic of trans-PKSs (Figure 3).[1]
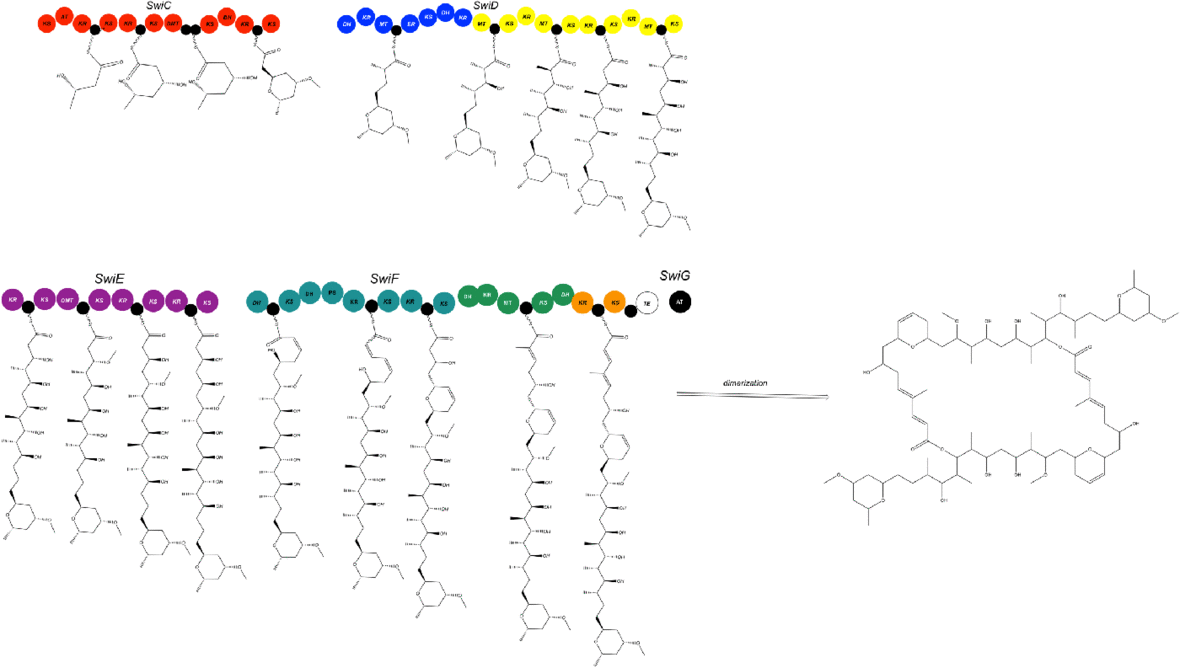
The swinholide biosynthesis gene cluster codes for a trans-AT PKS and does not integrate AT domains similar to the phormidolide (phm), miskinolide (mis), tolytoxin (tto), luminaolide (lum), and nosperin (nsp) gene clusters.[1] The swinholide biosynthesis gene cluster is also similar to the tto and lum gene clusters.[1] The swi and mis clusters both include four large genes encoding PKS enzymes and a gene encoding for the AT protein, but the order of the genes differs (Figure 4).[1]
In the swi biosynthesis gene cluster, first gene, SwiC, is on the reverse strand and the other four genes are facing the forward direction.[1] In the mis biosynthesis gene cluster, all genes are oriented in the same direction.[1] Although this is different, both swi and mis biosynthesis gene clusters are composed of similar catalytic domains.[1]
One distinct characteristic of the swi biosynthesis enzymes are its domain order, nonelongating domains, and split modules.[1] These are common features found in trans-PKSs.[1] There are four nonelongating ketosynthases in the swi cluster that are not a factor of the polyketide chain synthesis.[1] Three ketosynthases function to bind with modification enzymes and the fourth ketosynthase is found in the terminal section of SwiF.[1] There are only minor differences between swi and mis: two acyl carrier proteins (ACPs) in the middle of the SwiC protein, instead of a single ACP in the found in the MisC protein (Figure 4).[1]

In their monomeric structures, swi and mis have two varying ring structures.[1] In SwiF, the second and third dehydrotases (DHs) are located side-by-side (Figure 4).[1] For mis, the same DH-like domains were identified, but the third DH is pyran synthase (PS), which creates the dihydropyran ring in the structure of mis.[1] Further investigation revealed third DH in swi was a PS (Figure 3 and 5).[1] The other ring formation in mis was hypothesized to be catalyzed by either accessory enzymes or DH in MisC.[1] MisC and SwiC code for similar, but different DHs, but lack an overall PS domain in the dihydropyran ring formation.[1] The DH domains from MisC and SwiC were found to lack the glycine in a specific motif.[1] This, therefore, could indicate that a varying DH domain plays a vital role in ring formation.[1]
Despite the structural differences between swi and mis, the sequence identities of the genes differed from 73 to 85% even though there are structural similarities.[1] Scytophycin, tolytoxin, and luminaolide biosynthesis cluster genes also encompassed high sequence identity to swi and mis.[1] Although there is high sequence identity, SwiC and MisC proteins differ from alternative gene clusters, as shown in their chemical structures (Figure 1).[1]
Phylogenic analysis
The structural variants of the swinholide biosynthesis gene clusters origins were elucidate through phylogenic studies.[1] A phylogenetic tree of trans-encoded AT proteins showed that all six biosynthesis gene clusters were similar and assembled their own group.[1] Scytophycin, luminaolide, and tolytoxin biosynthesis gene clusters were arranged together based on ketosynthase domains, and the misakinolide and swinholide biosynthesis gene clusters constituted their own category (Figure 5).[1]

References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 1.25 1.26 1.27 1.28 1.29 1.30 1.31 1.32 1.33 1.34 1.35 1.36 "Nostoc sp. UHCC 0450". Applied and Environmental Microbiology 84 (3): e02321–17. November 2017. doi:10.1128/AEM.02321-17. PMID 29150506.
- ↑ Carmely, Shmuel; Kashman, Yoel (January 1985). "Structure of swinholide-a, a new macrolide from the marine sponge". Tetrahedron Letters 26 (4): 511–514. doi:10.1016/s0040-4039(00)61925-1.
- ↑ Kobayashi, Motomasa; Tanaka, Jun-ichi; Katori, Taketo; Matsuura, Miki; Kitagawa, Isao (January 1989). "Structure of swinholide A, a potent cytotoxic macrolide from the Okinawan marine sponge". Tetrahedron Letters 30 (22): 2963–2966. doi:10.1016/s0040-4039(00)99170-6.
- ↑ 4.0 4.1 Kitagawa, Isao; Kobayashi, Motomasa; Katori, Taketo; Yamashita, Megumi; Tanaka, Junichi; Doi, Mitsunobu; Ishida, Toshimasa (April 1990). "Absolute stereostructure of swinholide A, a potent cytotoxic macrolide from the Okinawan marine sponge Theonella swinhoei". Journal of the American Chemical Society 112 (9): 3710–2. doi:10.1021/ja00165a094.
- ↑ Sakai, Ryuichi; Higa, Tatsuo; Kashman, Yoel (September 1986). "Misakinolide-A, an Antitumor Macrolide from the Marine Sponge Theonella Sp.". Chemistry Letters 15 (9): 1499–1502. doi:10.1246/cl.1986.1499.
- ↑ 6.0 6.1 6.2 "Isolation of swinholide A and related glycosylated derivatives from two field collections of marine cyanobacteria". Organic Letters 7 (7): 1375–8. March 2005. doi:10.1021/ol050188x. PMID 15787510.
- ↑ "Hurghadolide A and swinholide I, potent actin-microfilament disrupters from the Red Sea sponge Theonella swinhoei". Journal of Natural Products 69 (1): 154–7. January 2006. doi:10.1021/np050404a. PMID 16441091.
- ↑ Tsukamoto, Sachiko; Ishibashi, Masami; Sasaki, Takuma; Kobayashi, Jun'Ichi (1991). "New congeners of swinholides from the okinawan marine sponge Theonella sp.". Journal of the Chemical Society, Perkin Transactions 1 (12): 3185–8. doi:10.1039/p19910003185.
- ↑ "Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts". Experientia 52 (7): 716–22. July 1996. doi:10.1007/bf01925581. PMID 8698116.
- ↑ "Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms" (in En). Nature Chemical Biology 11 (9): 705–12. September 2015. doi:10.1038/nchembio.1870. PMID 26236936.
 |
