Earth:Plastic degradation by marine bacteria
Plastic degradation in marine bacteria describes when certain pelagic bacteria break down polymers and use them as a primary source of carbon for energy. Polymers such as polyethylene(PE), polypropylene (PP), and polyethylene terephthalate (PET) are incredibly useful for their durability and relatively low cost of production, however it is their persistence and difficulty to be properly disposed of that is leading to pollution of the environment and disruption of natural processes. It is estimated that each year there are 9-14 million metric tons of plastic that are entering the ocean due to inefficient solutions for their disposal.[1] The biochemical pathways that allow for certain microbes to break down these polymers into less harmful byproducts has been a topic of study to develop a suitable anti-pollutant.[1]
Adaptive pressures
With the increasing presence of plastics in the environment, certain species of bacteria have evolved to degrade plastics into harmless by-products. Over the last 70 years, microbes have evolved to degrade plastics, as the global production of plastics steadily increased from 2 million metric tons to 380 million metric tons per year.[2] A study performed in 2021, led by Jan Zrimec of National Institute of Biology, Slovenia, was able to isolate 30,000 non-redundant enzyme homologues from more than 200 million genes in DNA samples obtained from the environment capable of degrading 10 different types of plastics.[1]
The results have showcased the impact of plastic pollution on the microbial environment and the tendency to adapt to a rapidly changing situation. A strong correlation can be seen between the microbial potential to degrade plastics and the content of global plastic pollution. Of the 30,000 enzyme homologues isolated, 12,000 were found in samples from the ocean.[1] Region specific analyses show that plastic degrading enzymes were found in high concentrations in deeper areas of the ocean where plastic pollution was more common.[1]
Metabolism
Diversity
With over 5000 grades of plastic polymers and variations in coatings such as flame retardants and pigments, diverse plastic polymer substrates suggest the existence of very heterogenous metabolic processes in plastic degradation.[3] Dynamic ocean conditions ranging in humidity, temperature, UV irradiation, pH, wind, and waves,[4] create varied growth conditions for bacteria and increase the possibility of diversified plastic degradation metabolisms.[3]
Mechanisms
As a developing topic, few studies have characterized the metabolic and biochemical mechanisms involved in the degradation of plastic by marine microbes.[5][6][7] A limited number of plastic degradation pathways in marine microbes have been extensively studied. It is important to note that although several metabolic processes in plastic degradation have been well-documented, these processes are likely not representative of the microbial population capable of plastic degradation.[3] Additionally, reaction times of plastic biodegradation metabolisms are poorly understood and are estimated to range between 1–400 hours in the marine environment.[4]
Polyethylene (PE)
Bacteria capable of polyethylene degradation have been described to utilize oxygenase to initiate biodegradation.[8] The formation of alcohol groups through oxygenase makes polyethylene more labile for degradation.[8] The hydrophilic properties of polyethylene polymers increase as the material experiences degradation and oxidation, which causes polyethylene to become less recalcitrant.[3] Lipases, esterase, endopeptidases, and other extracellular enzymes then further degrade the polyethylene polymers.[3] The role of laccase in polyethylene degradation by Rhodococcus ruber is well-documented as an important enzyme for biodegradation.[9] Alkane hydroxylase is thought to play a similar role in pseudomonas species capable of polyethylene degradation.[10] Once enzymes degrade polyethylene polymers into oligomers, microbial cells uptake the molecules through either Major Facilitator Superfamily proteins or ATP binding cassettes.[10] The polyethylene oligomers are converted into Acetyl-CoA and succinyl-CoA and enter the tricarboxylic acid cycle, and eventually the respiratory chain to produce ATP.[3]
Polyethylene Terephthalate (PET)
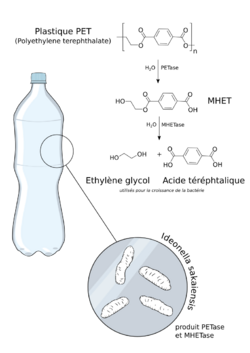
Aromatic rings in the structure of polyethylene terephthalate pose challenges for microbial biodegradation.[6] Despite the challenge of degrading aromatic rings, several microbes are documented to use polyethylene terephthalate as a sole energy and carbon source.[3] Microbes that utilize PET degradation first adhere to the substrate surface and release enzymes such as hydrolases and cutinases.[11] Following the activity of enzymes like MHETase, molecules from PET degradation are taken up by active transport into bacterial cells.[3][12] As demonstrated in Figure 2, transported PET molecules consist of terephthalic acid and ethylene glycol, monomers of PET.[3] Certain microbes can use these monomers as their primary carbon source leading to PET degradation. The most relevant pathways related to the degradation of the monomer ethylene glycol are the acetaldehyde/ethanol pathway and the glyoxylic acid pathway. The glyoxylic acid pathway is commonly studied in the Pseudomonas genus as many species such as P. putida are known to be highly capable of degrading polyethylene in aquatic environments.[13] Following a series of chemical processes, the terephthalic acid is converted into 4-carboxy-2-hydroxymuconic, which is a precursor molecule to the TCA cycle and eventually converted into pyruvate and oxaloacetate.[3] Several bacterial species in Betaproteobacteria, Myxococcota (formerly included in Deltaproteobacteria), and Gammaproteobacteria are capable of PET Biodegradation.[14]
Polystyrene (PS)
Polystyrene consists of molecules with both strong hydrophobicity and a high molecular weight.[3] Bacteria that are capable of degrading this molecule are documented to release monooxygenases to initiate the oxidization of polystyrene molecules.[3][15] Following the monooxygenase step, the polystyrene molecule is transformed into phenylacetic acid during the upper pathway of styrene metabolism.[16] Phenylacetic acid is first converted into phenylacetyl-coA, and later acetyl-CoA and succinyl-CoA after a series of enzymatic reactions.[3] Acetyl-CoA and succinyl-CoA then enter the tricarboxylic acid cycle.[16] Several Rhodococcus ruber strains are capable of polystyrene biodegradation.[17]
Polyhydroxyalkanoate (PHA)
Polyhydroxyalkanoate (PHA) is a polyester that is naturally produced by various microorganisms in response to nutrient limitation and environmental stress.[15] Although polyhydroxyalkanoates have a microbial origin and are often exploited for commercial use,[18] the polymer is also biodegraded by several species of bacteria.[15][19] Polyhydroxyalkanoate biodegradation is reported to occur in various environments including marine habitats.[3] The biodegradation process of polyhydroxyalkanoate varies between bacterial species that both produce and degrade polyhydroxyalkanoate, and species that only degrade polyhydroxyalkanoate.[3] Species that cannot produce polyhydroxyalkanoate but are capable of polyhydroxyalkanoate degradation initiate hydrolysis of the substrate externally with ectoenzymes that yield hydroxybutyrate.[20] The resulting oligomers cross bacterial membranes through passive diffusion in a form that can directly enter β-oxidation to yield acetyl-CoA.[3] The resulting acetyl-CoA then enters the TCA cycle.[3] Several bacterial species in the genera Gracilibacillus, Enterobacter, and Bacillus are capable of polyhydroxyalkanoate biodegradation.[19]
Challenges
Literature frequently discusses the biological constraints that organisms must overcome to degrade plastic.[21][5] Features that make plastic challenging to degrade include long-chain polymers, high molecular weight, hydrophobicity, and crystallinity.[6] Although hydrocarbons found in plastic are potential sources of carbon and energy for bacteria, the lack of essential nutrients like nitrogen in plastic make it insufficient to support microbial growth without additional nutrient sources.[22]
Species
| Bacteria | Type of plastic | Characteristics |
|---|---|---|
| Kocuria palustris M16 | PE[23] | After 30 days, weight loss of polyethylene was 1% after incubation with Kocuria palustris M16 [23] |
| Rhodococcus ruber | PE [24] | Found ubiquitously in many different habitats. Degraded up to 8% of polyolefin within 30 days of incubation.[25] Formed a dense biofilm on the surface of polyethylene films and used polyolefin as carbon and energy source.[24] |
| Ideonella sakaiensis | PET[26] | Idonella sakaiensis was discovered in a PET recycling site. It was using PET film as a major carbon source for growth. The bacterium was part of a consortium that degraded PET film at a rate of 0.13 mg cm−2 day−1 and had nearly destroyed a PET film in 6 weeks.[26] |
| Marinobacter sp. | LDPE[27] | Many different species have been identified as being involved in hydrocarbon degradation.[28] Most prominently, H-246 was incubated with low-density polyethylene film for 90 days and showed a maximum weight loss of 1.68%.[27] |
| Exiguobacterium sp., Halomonas sp. and Ochrobactrum sp. | PE and PET [29] | A marine bacterial community collected from the upper tidal area in Huiquan Bay consisting of Exiguobacterium sp., Halomonas sp. and Ochrobactrum sp. was shown to degrade PET and PE.[29] |
| Bacillus pumilus M27 | PE [23] | After 30 days, weight loss of polyethylene was 1.5% after incubation with Bacillus pumilus M27 [23] |
| Bacillus subtilis H1584 | PE [23] | After 30 days, weight loss of polyethylene was 1.75% after incubation with Bacillus subtilis H1584 [23] |
| Phormidium | PET,[30] PP, PE [31] | A filamentous cyanobacterium collected in the North Sea was found to be colonizing PET bottles and capable of degrading hydrocarbons.[30][31] |
| Lewinella | PET [30] | A bacterium found in the North Sea colonizing PET bottles.[30][32] |
| Rivularia | PP, PE [31] | Found degrading PP and PE microplastics in the North Atlantic.[31][32] |
| Stanieria | PET [33] | A cyanobacteria that was found to be abundant on PET across stations in the summer season in the North Sea, the English Channel, the Celtic Sea and the Bristol Channel.[33][32] |
| Pseudomonadota, Bacteroides | Microplastic[34] | These bacteria were found degrading microplastics in the Belgian part of the North Sea.[34][32] |
| Shewanella, Moritella, Psychrobacter | PCL [35] | These microorganisms were found degrading PCL in deep-sea sediment in the Kurile and Japan Trenches.[35][32] |
| Pseudomonas sp. | PCL,[35] Monofilament fibres of PCL, PHB/V, PBS [36] | Pseudomonas was found degrading PCL in deep-sea sediment in the Kurile and Japan Trenches.[35][32] It was also found in deep seawater from Rausu and Toyama, Japan degrading monofilaments of PCL, PHB/v, and PBS in the temperature range of 4-25 degrees Celsius.[36] |
| Vibrio alginolyticus | PVA-LLDPE [37] | Strains BTTC10 and BTTC27 collected from benthic zones of different marine environments were shown to decrease the tensile strength of plastic films combining PVA and LLDPE plastic.[37] |
| Vibrio parahaemolyticus | PVA-LLDPE [37] | Strains BTTV4 and BTTN18 collected from benthic zones of different marine environments were shown to decrease the tensile strength of plastic films combining PVA and LLDPE plastic.[37] |
| Alcanivorax sp. | Monofilament fibers of PCL, PHB/V, PBS [36] | This bacterial strain was collected from Kume Island in Japan. It was found to feed on monofilaments of PCL, PHB/V, PBS at 25 degrees Celsius.[36] |
| Tenacibaculum sp. | Monofilament fibers of PCL, PHB/V, PBS [36] | Bacteria belonging to this genus was found in Rausu, Toyama, and Kum Japan. It is able to degrade PCL fibers in vitro at low temperatures (4-10 degrees Celsius).[36] |
Impact

According to the National Ocean Service, it is estimated that there are 8 million metric tons of plastic in the ocean.[40] Ocean plastic affects many marine species in the form of whole plastic and micro plastics. Since the discovery of bacteria that can feed on plastic, there has been hope that these microbes could help clean the ocean of plastic, but Ramani Narayan, a professor in chemical engineering at Michigan State University says that this viewpoint misses the point.[41] Moreover, after Kale et al. performed an extensive review of all data available on these bacteria, they have found that there are currently no practical industrial applications of these microbes in environments to make a substantial impact on the plastic problem in the ocean.[42] This can be attributed to findings that have found the rate of degradation by these microbes to be low, even when optimized in laboratory settings.[43] Hence, researchers at the University of Portsmouth have been working on genetically engineering these bacteria to be more efficient at degrading the plastic[citation needed].
Genetic engineering
Scientists are working on genetically engineering Ideonella sakaiensis to break down PET plastic at a faster rate in order to make it a viable option to help recycle plastic.[44] Researchers at the University of Portsmouth have discovered that mixing PETase with a second enzyme called MHETase has created an “enzyme cocktail” that degrades PET plastic at 6 times the rate it did before.[45][46] In an interview done by Bloomberg QuickTake, Professor John McGeehan, who worked with the team at the University of Portsmouth to make this discovery, spoke of the possibility to create an enzyme powder that could be distributed across plastic recycling facilities to break down plastics to their constituent components, Ethylene Glycol and Terephthalic Acid.[47] These building blocks could then be used to make new plastics, which would create a more efficient and effective way of recycling plastics. However, Dr. Hermann J. Heipieper from the Helmholtz Center for Environmental Research does not see this as a viable option due to the high bond strength found in plastic.[47]
Another major part of the research being done on microbial environmental remediation involves manipulating the genomes of specific microbes to increase their metabolic power. For example, in Pseudomonas putida, scientists have engineered a strain to overexpress glc and glcDEF, operons that contain genes coding for enzymes that convert toxic intermediates into the next substrates in the pathway. This would allow plastic monomers such as ethylene glycol to be converted into high-value chemicals such as medium-chain-length polyhydroxyalkanoates (mcl-PHA). As a natural bioplastic, PHAs have similar properties to synthetic plastics and are biodegradable as well as nontoxic making them useful for biomedical applications. Its utility even extends to its potential for the production of hydrocarbon jet fuel if further chemically catalyzed.[48]
Genetically engineered bacteria also do not have a practical application in the ocean, yet, according to the Ocean Conservatory group.[49] Dr. Naryan believes that releasing genetically engineered bacteria into the ocean ecosystems could be irresponsible and have many negative side effects on the ecosystem.[41] As the Ocean Conservatory group states, the solution to the plastic problem does not lie in genetically engineered bacteria, but rather decreasing the plastic input into the ocean and increasing collection and recycling efforts.[49]
With these potential limitations in mind, a deeper comparison of the pros and cons of engineering microbes for plastic degradation should be conducted. As mentioned, one problem in environmental remediation of plastics is that only 30% of plastics are collected to be recycled.[50] Without efficient collection in the first place, the high energy cost of enzymatic recycling would be more burdening than the amount of waste it would break down. Another problem includes the fact that this may not be applicable at a wide scale since enzymes have a short half-life so engineered organisms may not remain catalytically active for long enough to be effective. On the other hand, to fix unpredictable enzyme-polymer interactions, there has been development of new techniques such as computational tools to visualize the 3-D interactions between plastics and enzymes.[51] Thus, engineered microbes can remain as a hopeful solution for plastic remediation.
Toxicity of degradation products
A study was performed to understand the toxicity of the degradation of polythene bags and cups by P. aeruginosa, Streptomyces sp., Aspergillus niger, Staphylococcus aureus, and Rhizopus sp.[53] The study found carbon dioxide gas to be the main byproduct of the degradation process of polythene by the bacteria mentioned before. However, the particles produced as a byproduct of PE bio-degradation had negative impacts on the production of polysaccharides, proteins, and nutrient uptake in roots of plants.[53] Another study performed by Aswale focused on how biodegraded polythene affected seed germination in plants. It found that biodegraded polythene was correlated with a decrease in percentage of seed germination, indicating that the byproducts of the breakdown could have negative effects on the seed health.[54]
Food web biomagnification
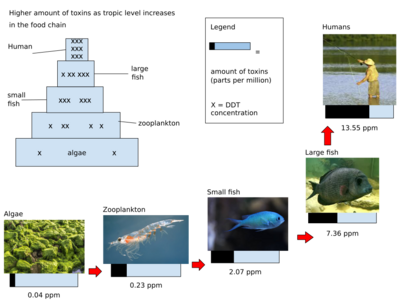
As seen in Figure 4, food web biomagnification refers to the process by which concentrations of contaminants increase as the trophic levels increase.[55] In terms of plastic, this means when plankton eat plastic, then fish eat the plankton, and larger fish eat that fish, the amount of plastic accumulates in the largest fish.[55] Therefore, removing plastic from the system at the bacteria level would prevent the plastic from bioaccumulating in larger fish. However, as mentioned above, these bacteria are not very efficient at degrading plastic, and therefore do not have the capabilities to create a substantial impact on this problem.[43]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Zrimec, Jan; Kokina, Mariia; Jonasson, Sara; Zorrilla, Francisco; Zelezniak, Aleksej (2021-10-26). "Plastic-Degrading Potential across the Global Microbiome Correlates with Recent Pollution Trends". mBio 12 (5): e0215521. doi:10.1128/mbio.02155-21. ISSN 2150-7511. PMID 34700384.
- ↑ Geyer, Roland (19 Jul 2017). "Production, use, and fate of all plastics ever made". Science Advances 3 (7): 7. doi:10.1126/sciadv.1700782. PMID 28776036. Bibcode: 2017SciA....3E0782G.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 Jacquin, Justine; Cheng, Jingguang; Odobel, Charlène; Pandin, Caroline; Conan, Pascal; Pujo-Pay, Mireille; Barbe, Valérie; Meistertzheim, Anne-Leila et al. (2019). "Microbial Ecotoxicology of Marine Plastic Debris: A Review on Colonization and Biodegradation by the "Plastisphere"". Frontiers in Microbiology 10: 865. doi:10.3389/fmicb.2019.00865. ISSN 1664-302X. PMID 31073297.
- ↑ 4.0 4.1 Oliveira, Juliana; Belchior, Afonso; da Silva, Verônica D.; Rotter, Ana; Petrovski, Željko; Almeida, Pedro L.; Lourenço, Nídia D.; Gaudêncio, Susana P. (2020). "Marine Environmental Plastic Pollution: Mitigation by Microorganism Degradation and Recycling Valorization". Frontiers in Marine Science 7. doi:10.3389/fmars.2020.567126. ISSN 2296-7745.
- ↑ 5.0 5.1 Singh, Baljit; Sharma, Nisha (2008-03-01). "Mechanistic implications of plastic degradation" (in en). Polymer Degradation and Stability 93 (3): 561–584. doi:10.1016/j.polymdegradstab.2007.11.008. ISSN 0141-3910. https://www.sciencedirect.com/science/article/pii/S0141391007003539.
- ↑ 6.0 6.1 6.2 Urbanek, Aneta K.; Rymowicz, Waldemar; Mirończuk, Aleksandra M. (2018). "Degradation of plastics and plastic-degrading bacteria in cold marine habitats". Applied Microbiology and Biotechnology 102 (18): 7669–7678. doi:10.1007/s00253-018-9195-y. ISSN 0175-7598. PMID 29992436.
- ↑ Kolvenbach, Boris A; Helbling, Damian E; Kohler, Hans-Peter E; Corvini, Philippe F-X (2014-06-01). "Emerging chemicals and the evolution of biodegradation capacities and pathways in bacteria" (in en). Current Opinion in Biotechnology. Energy biotechnology • Environmental biotechnology 27: 8–14. doi:10.1016/j.copbio.2013.08.017. ISSN 0958-1669. PMID 24863891. https://www.sciencedirect.com/science/article/pii/S0958166913006411.
- ↑ 8.0 8.1 Krueger, Martin C.; Harms, Hauke; Schlosser, Dietmar (2015). "Prospects for microbiological solutions to environmental pollution with plastics". Applied Microbiology and Biotechnology 99 (21): 8857–8874. doi:10.1007/s00253-015-6879-4. ISSN 1432-0614. PMID 26318446. https://pubmed.ncbi.nlm.nih.gov/26318446/.
- ↑ Santo, Miriam; Weitsman, Ronen; Sivan, Alex (2013-10-01). "The role of the copper-binding enzyme – laccase – in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber" (in en). International Biodeterioration & Biodegradation 84: 204–210. doi:10.1016/j.ibiod.2012.03.001. ISSN 0964-8305. https://www.sciencedirect.com/science/article/pii/S0964830512000637.
- ↑ 10.0 10.1 Gravouil, Kévin; Ferru-Clément, Romain; Colas, Steven; Helye, Reynald; Kadri, Linette; Bourdeau, Ludivine; Moumen, Bouziane; Mercier, Anne et al. (2017-05-02). "Transcriptomics and Lipidomics of the Environmental Strain Rhodococcus ruber Point out Consumption Pathways and Potential Metabolic Bottlenecks for Polyethylene Degradation". Environmental Science & Technology 51 (9): 5172–5181. doi:10.1021/acs.est.7b00846. ISSN 1520-5851. PMID 28345896. Bibcode: 2017EnST...51.5172G. https://pubmed.ncbi.nlm.nih.gov/28345896/.
- ↑ Danso, Dominik; Schmeisser, Christel; Chow, Jennifer; Zimmermann, Wolfgang; Wei, Ren; Leggewie, Christian; Li, Xiangzhen; Hazen, Terry et al. (2018-04-15). "New Insights into the Function and Global Distribution of Polyethylene Terephthalate (PET)-Degrading Bacteria and Enzymes in Marine and Terrestrial Metagenomes". Applied and Environmental Microbiology 84 (8): e02773–17. doi:10.1128/AEM.02773-17. ISSN 1098-5336. PMID 29427431. Bibcode: 2018ApEnM..84E2773D.
- ↑ Hosaka, Masaru; Kamimura, Naofumi; Toribami, Shotaro; Mori, Kosuke; Kasai, Daisuke; Fukuda, Masao; Masai, Eiji (2013). "Novel Tripartite Aromatic Acid Transporter Essential for Terephthalate Uptake in Comamonas sp. Strain E6". Applied and Environmental Microbiology 79 (19): 6148–6155. doi:10.1128/AEM.01600-13. ISSN 0099-2240. PMID 23913423. Bibcode: 2013ApEnM..79.6148H.
- ↑ Mückschel, Björn; Simon, Oliver; Klebensberger, Janosch; Graf, Nadja; Rosche, Bettina; Altenbuchner, Josef; Pfannstiel, Jens; Huber, Armin et al. (2012-12-15). "Ethylene Glycol Metabolism by Pseudomonas putida" (in en). Applied and Environmental Microbiology 78 (24): 8531–8539. doi:10.1128/AEM.02062-12. ISSN 0099-2240. PMID 23023748.
- ↑ Danso, Dominik; Schmeisser, Christel; Chow, Jennifer; Zimmermann, Wolfgang; Wei, Ren; Leggewie, Christian; Li, Xiangzhen; Hazen, Terry et al. (2018-04-15). Parales, Rebecca E.. ed. "New Insights into the Function and Global Distribution of Polyethylene Terephthalate (PET)-Degrading Bacteria and Enzymes in Marine and Terrestrial Metagenomes" (in en). Applied and Environmental Microbiology 84 (8): e02773–17. doi:10.1128/AEM.02773-17. ISSN 0099-2240. PMID 29427431. Bibcode: 2018ApEnM..84E2773D.
- ↑ 15.0 15.1 15.2 Pathak, Vinay Mohan; Navneet (2017-03-23). "Review on the current status of polymer degradation: a microbial approach". Bioresources and Bioprocessing 4 (1): 15. doi:10.1186/s40643-017-0145-9. ISSN 2197-4365.
- ↑ 16.0 16.1 Luu, Rita A.; Schneider, Benjamin J.; Ho, Christie C.; Nesteryuk, Vasyl; Ngwesse, Stacy E.; Liu, Xianxian; Parales, Juanito V.; Ditty, Jayna L. et al. (2013). "Taxis of Pseudomonas putida F1 toward Phenylacetic Acid Is Mediated by the Energy Taxis Receptor Aer2". Applied and Environmental Microbiology 79 (7): 2416–2423. doi:10.1128/AEM.03895-12. ISSN 0099-2240. PMID 23377939. Bibcode: 2013ApEnM..79.2416L.
- ↑ Mor, Roi; Sivan, Alex (2008-11-01). "Biofilm formation and partial biodegradation of polystyrene by the actinomycete Rhodococcus ruber" (in en). Biodegradation 19 (6): 851–858. doi:10.1007/s10532-008-9188-0. ISSN 1572-9729. PMID 18401686. https://doi.org/10.1007/s10532-008-9188-0.
- ↑ Ray, Subhasree; Kalia, Vipin Chandra (2017). "Biomedical Applications of Polyhydroxyalkanoates". Indian Journal of Microbiology 57 (3): 261–269. doi:10.1007/s12088-017-0651-7. ISSN 0046-8991. PMID 28904409.
- ↑ 19.0 19.1 Volova, T. G.; Boyandin, A. N.; Vasiliev, A. D.; Karpov, V. A.; Prudnikova, S. V.; Mishukova, O. V.; Boyarskikh, U. A.; Filipenko, M. L. et al. (2010-12-01). "Biodegradation of polyhydroxyalkanoates (PHAs) in tropical coastal waters and identification of PHA-degrading bacteria" (in en). Polymer Degradation and Stability 95 (12): 2350–2359. doi:10.1016/j.polymdegradstab.2010.08.023. ISSN 0141-3910. https://www.sciencedirect.com/science/article/pii/S0141391010003642.
- ↑ O'Leary, Niall D.; O'Connor, Kevin E.; Ward, Patrick; Goff, Miriam; Dobson, Alan D. W. (2005). "Genetic Characterization of Accumulation of Polyhydroxyalkanoate from Styrene in Pseudomonas putida CA-3". Applied and Environmental Microbiology 71 (8): 4380–4387. doi:10.1128/AEM.71.8.4380-4387.2005. ISSN 0099-2240. PMID 16085828. Bibcode: 2005ApEnM..71.4380O.
- ↑ Chiellini, E.; Corti, A.; D'Antone, S.; Baciu, R. (2006-11-01). "Oxo-biodegradable carbon backbone polymers – Oxidative degradation of polyethylene under accelerated test conditions" (in en). Polymer Degradation and Stability 91 (11): 2739–2747. doi:10.1016/j.polymdegradstab.2006.03.022. ISSN 0141-3910. https://www.sciencedirect.com/science/article/pii/S0141391006001509.
- ↑ Sauret, Caroline; Tedetti, Marc; Guigue, Catherine; Dumas, Chloé; Lami, Raphaël; Pujo-Pay, Mireille; Conan, Pascal; Goutx, Madeleine et al. (2016). "Influence of PAHs among other coastal environmental variables on total and PAH-degrading bacterial communities". Environmental Science and Pollution Research International 23 (5): 4242–4256. doi:10.1007/s11356-015-4768-0. ISSN 1614-7499. PMID 26122564. https://pubmed.ncbi.nlm.nih.gov/26122564/.
- ↑ 23.0 23.1 23.2 23.3 23.4 23.5 Harshvardhan, Kumar; Jha, Bhavanath (7 Nov 2013). "Biodegradation of low-density polyethylene by marine bacteria from pelagic waters, Arabian Sea, India". Marine Pollution Bulletin 77 (1–2): 100–106. doi:10.1016/j.marpolbul.2013.10.025. PMID 24210946. https://pubmed.ncbi.nlm.nih.gov/24210946/.
- ↑ 24.0 24.1 Gilan, Irit; Sivan, Alex (18 Mar 2013). "Effect of proteases on biofilm formation of the plastic-degrading actinomycete Rhodococcus ruber C208". FEMS Microbiology Letters 342 (1): 18–23. doi:10.1111/1574-6968.12114. PMID 23448092.
- ↑ Gilan Orr, I; Hadar, Y; Sivan, A (19 Feb 2004). "Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber". Applied Microbiology and Biotechnology 65 (1): 97–104. doi:10.1007/s00253-004-1584-8. PMID 15221232. https://pubmed.ncbi.nlm.nih.gov/15221232/.
- ↑ 26.0 26.1 Yoshida, Shosuke; Hiraga, Kazumi; Takehana, Toshihiko; Taniguchi, Ikuo; Yamaji, Hironao; Maeda, Yasuhito; Toyohara, Kiyotsuna; Miyamoto, Kenji et al. (2016). "A bacterium that degrades and assimilates poly(ethylene terephthalate)". Science 351 (6278): 1196–1199. doi:10.1126/science.aad6359. ISSN 0036-8075. PMID 26965627. Bibcode: 2016Sci...351.1196Y. https://www.jstor.org/stable/24743191.
- ↑ 27.0 27.1 Khandare, Shrikant; Chaudhary, Doongar; Jha, Bhavanath (5 Feb 2021). "Marine bacterial biodegradation of low-density polyethylene (LDPE) plastic". Biodegradation 32 (2): 127–143. doi:10.1007/s10532-021-09927-0. PMID 33544248. https://pubmed.ncbi.nlm.nih.gov/33544248/.
- ↑ Yakimov, Michail; Timmis, Kenneth; Golyshin, Peter (9 May 2007). "Obligate oil-degrading marine bacteria". Current Opinion in Biotechnology 18 (3): 257–66. doi:10.1016/j.copbio.2007.04.006. PMID 17493798. https://pubmed.ncbi.nlm.nih.gov/17493798/.
- ↑ 29.0 29.1 Gao, Rongrong; Sun, Chaomin (24 Apr 2021). "A marine bacterial community capable of degrading poly(ethylene terephthalate) and polyethylene". Journal of Hazardous Materials 416: 125928. doi:10.1016/j.jhazmat.2021.125928. PMID 34489083.
- ↑ 30.0 30.1 30.2 30.3 Oberbeckmann, Sonja; Osborn, A Mark; Duhaime, Melissa (3 Aug 2016). "Microbes on a Bottle: Substrate, Season and Geography Influence Community Composition of Microbes Colonizing Marine Plastic Debris". PLOS ONE 11 (8): e0159289. doi:10.1371/journal.pone.0159289. PMID 27487037. Bibcode: 2016PLoSO..1159289O.
- ↑ 31.0 31.1 31.2 31.3 Zettler, Erik; Mincer, Tracy; Amaral-Zettler, Linda (2013). "Life in the "Plastisphere": Microbial Communities on Plastic Marine Debris". Environ. Sci. Technol. 47 (13): 7137–7146. doi:10.1021/es401288x. PMID 23745679. Bibcode: 2013EnST...47.7137Z. https://pubs.acs.org/doi/10.1021/es401288x.
- ↑ 32.0 32.1 32.2 32.3 32.4 32.5 Urbanek, Aneta; Rymowicz, Waldemar; Mironczuk, Aleksandra (2018). "Degradation of plastics and plastic-degrading bacteria in cold marine habitats". Applied Microbiology and Biotechnology 102 (18): 7669–7678. doi:10.1007/s00253-018-9195-y. PMID 29992436.
- ↑ 33.0 33.1 Oberbeckmann, Sonja; Leoder, Martin; Gerdts, Gunnar; Osborn, A Mark (Nov 2014). "Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters". FEMS Microbiology Ecology 90 (2): 478–492. doi:10.1111/1574-6941.12409. PMID 25109340.
- ↑ 34.0 34.1 Tender, Caroline; Devriese, Lisa; Haegeman, Annelies; Maes, Sara; Ruttink, Tom; Dawyndt, Peter (18 Aug 2015). "Bacterial Community Profiling of Plastic Litter in the Belgian Part of the North Sea". Environmental Science & Technology 49 (16): 9629–38. doi:10.1021/acs.est.5b01093. PMID 26204244. Bibcode: 2015EnST...49.9629D. https://pubmed.ncbi.nlm.nih.gov/26204244/.
- ↑ 35.0 35.1 35.2 35.3 Sekiguchi, Takayoshi; Sato, Takako; Enoki, Makiko; Kanehiro, Haruyuki (Jan 2011). "Isolation and characterization of biodegradable plastic degrading bacteria from deep-sea environments". JAMSTEC Report of Research and Development 11: 33–41. doi:10.5918/jamstecr.11.33. https://www.researchgate.net/publication/273991590.
- ↑ 36.0 36.1 36.2 36.3 36.4 36.5 Sekiguchi, Takayoshi; Saika, Azusa; Nomura, Koji; Watanabe, Toshihiro; Wantanabe, Toru; Fujimonto, Yu; Enoki, Makiko; Sato, Takako et al. (Jul 2011). "Biodegradation of aliphatic polyesters soaked in deep seawaters and isolation of poly(ɛ-caprolactone)-degrading bacteria". Polymer Degradation and Stability 96 (6): 1397–1403. doi:10.1016/j.polymdegradstab.2011.03.004. https://www.sciencedirect.com/science/article/pii/S014139101100108X.
- ↑ 37.0 37.1 37.2 37.3 Sasidharan, Rahul; Bhat, Sarita; Muthusamy, Chandrasekaran; Francis, Vidya (May 2013). "Biodegradation of polyvinyl alcohol-low linear density polyethylene-blended plastic film by consortium of marine benthic vibrio". International Journal of Environmental Science and Technology 11 (7): 1827–1834. doi:10.1007/s13762-013-0335-8. https://www.researchgate.net/publication/256765453.
- ↑ Janatunaim, Rifqi Z.; Fibriani, Azzania (2020). "Construction and Cloning of Plastic-degrading Recombinant Enzymes (MHETase)". Recent Patents on Biotechnology 14 (3): 229–234. doi:10.2174/1872208314666200311104541. ISSN 2212-4012. PMID 32160855. https://pubmed.ncbi.nlm.nih.gov/32160855/.
- ↑ "pGLO Plasmid Map and Resources" (in en-us). https://www.bio-rad.com/en-us/applications-technologies/pglo-plasmid-map-resources?ID=NISQOC15.
- ↑ "A Guide to Plastic in the Ocean". National Ocean and Atmospheric Administration. https://oceanservice.noaa.gov/hazards/marinedebris/plastics-in-the-ocean.html.
- ↑ 41.0 41.1 "Plastic-eating Microbes Won't Solve Our Ocean Debris Problem" (in en). https://www.capeandislands.org/show/living-lab-radio-on-cai/2018-04-23/plastic-eating-microbes-wont-solve-our-ocean-debris-problem.
- ↑ Kale, Swapnil; Deshmukh, Amit; Dudhare, Mahendra; Patil, Vikram (4 Nov 2015). "Microbial degradation of plastic: a review". J Biochem Tech 6 (2): 952–961.
- ↑ 43.0 43.1 Krueger, Martin; Harms, Hauke; Schlosser, Dietmar (Nov 2015). "Prospects for microbiological solutions to environmental pollution with plastics". Applied Microbiology and Biotechnology 99 (21): 8857–74. doi:10.1007/s00253-015-6879-4. PMID 26318446. https://pubmed.ncbi.nlm.nih.gov/26318446/.
- ↑ Carpenter, Scott. "The Race To Develop Plastic-Eating Bacteria" (in en). https://www.forbes.com/sites/scottcarpenter/2021/03/10/the-race-to-develop-plastic-eating-bacteria/.
- ↑ "New Enzyme Cocktail Digests Plastic Waste Six Times Faster". University of Portsmouth. https://www.port.ac.uk/news-events-and-blogs/news/new-enzyme-cocktail-digests-plastic-waste-six-times-faster..
- ↑ Knott, Brandon; Erickson, Erika; Allen, Mark; Gado, Japheth; Graham, Rosie; Kearns, Fiona; Pardo, Isabel; Topuzlu, Ece et al. (2020). "Characterization and Engineering of a Two-Enzyme System for Plastics Depolymerization". Proceedings of the National Academy of Sciences 117 (41): 25476–25485. doi:10.1073/pnas.2006753117. PMID 32989159. Bibcode: 2020PNAS..11725476K.
- ↑ 47.0 47.1 Tom, Gibson. "Could Plastic-Eating Bacteria Save The Planet?". Bloomberg Quicktake. https://www.youtube.com/watch?v=DDhPuyrSq3E&t=207s&ab_channel=BloombergQuicktake.
- ↑ Franden, Mary Ann; Jayakody, Lahiru N.; Li, Wing-Jin; Wagner, Neil J.; Cleveland, Nicholas S.; Michener, William E.; Hauer, Bernhard; Blank, Lars M. et al. (July 2018). "Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization" (in en). Metabolic Engineering 48: 197–207. doi:10.1016/j.ymben.2018.06.003. PMID 29885475.
- ↑ 49.0 49.1 "Mutant Enzymes are Cool, But Not Likely to Solve Our Ocean Plastics Problem" (in en). 2018-04-19. https://oceanconservancy.org/blog/2018/04/19/mutant-enzymes-cool-not-likely-solve-ocean-plastics-problem/.
- ↑ Kaushal, Jyoti; Khatri, Madhu; Arya, Shailendra Kumar (June 2021). "Recent insight into enzymatic degradation of plastics prevalent in the environment: A mini - review" (in en). Cleaner Engineering and Technology 2: 100083. doi:10.1016/j.clet.2021.100083.
- ↑ Qi, Xinhua; Yan, Wenlong; Cao, Zhibei; Ding, Mingzhu; Yuan, Yingjin (2021-12-26). "Current Advances in the Biodegradation and Bioconversion of Polyethylene Terephthalate". Microorganisms 10 (1): 39. doi:10.3390/microorganisms10010039. ISSN 2076-2607. PMID 35056486.
- ↑ "Biomagnification - an overview". https://www.sciencedirect.com/topics/earth-and-planetary-sciences/biomagnification#:~:text=Biomagnification%20is%20the%20accumulation%20of,greater%20than%20expected%20from%20equilibrium.
- ↑ 53.0 53.1 Venkatesh, S.; Mahboob, Shahid; Govindarajan, Marimuthu; Al-Ghanim, Khalid A.; Ahmed, Zubair; Al-Mulhm, Norah; Gayathri, R.; Vijayalakshmi, S. (2021-05-01). "Microbial degradation of plastics: Sustainable approach to tackling environmental threats facing big cities of the future" (in en). Journal of King Saud University - Science 33 (3): 101362. doi:10.1016/j.jksus.2021.101362. ISSN 1018-3647.
- ↑ Aswale, Pranita Nandkumar (29 Jan 2011). Studies on biodegradation of polythene (PhD thesis). Dr. Babasaheb Ambedkar Marathwada University. hdl:10603/78820.
- ↑ 55.0 55.1 Hale, Robert. "Biomagnification". https://www.sciencedirect.com/topics/earth-and-planetary-sciences/biomagnification#:~:text=Biomagnification%20is%20the%20accumulation%20of,greater%20than%20expected%20from%20equilibrium.
 |