Biology:Streptomyces
| Streptomyces | |
|---|---|
| |
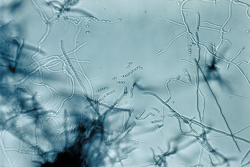
| Slide culture | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Actinomycetota |
| Class: | Actinomycetia |
| Order: | Streptomycetales |
| Family: | Streptomycetaceae |
| Genus: | Streptomyces Waksman and Henrici 1943 (Approved Lists 1980) |
| Type species | |
| Streptomyces albus (Rossi Doria 1891) Waksman and Henrici 1943
| |
| Diversity | |
| About 550 species | |
| Synonyms[1] | |
|
List
| |
Streptomyces is the largest genus of Actinomycetota, and the type genus of the family Streptomycetaceae.[3] Over 700 species of Streptomyces bacteria have been described.[4][5][6] As with the other Actinomycetota, streptomycetes are gram-positive, and have very large genomes with high GC content.[5][7] Found predominantly in soil and decaying vegetation, most streptomycetes produce spores, and are noted for their distinct "earthy" odor that results from production of a volatile metabolite, geosmin.[8] Different strains of the same species may colonize very diverse environments.[5]
Streptomycetes are characterised by a complex secondary metabolism.[7] Between 5-23% (average: 12%) of the protein-coding genes of each Streptomyces species are implicated in secondary metabolism.[5] Streptomycetes produce over two-thirds of the clinically useful antibiotics of natural origin (e.g., neomycin, streptomycin, cypemycin, grisemycin, bottromycins and chloramphenicol).[9][10] The antibiotic streptomycin takes its name directly from Streptomyces. Streptomycetes are infrequent pathogens, though infections in humans, such as mycetoma, can be caused by S. somaliensis and S. sudanensis, and in plants can be caused by S. caviscabies, S. acidiscabies, S. turgidiscabies and S. scabies.
Taxonomy
Streptomyces is the type genus of the family Streptomycetaceae[11] and currently covers more than 700 species with the number increasing every year.[12][6] It is estimated that the total number of Streptomyces species is close to 1600.[5] Acidophilic and acid-tolerant strains that were initially classified under this genus have later been moved to Kitasatospora (1997) [13] and Streptacidiphilus (2003).[14] Species nomenclature are usually based on their color of hyphae and spores.
Saccharopolyspora erythraea was formerly placed in this genus (as Streptomyces erythraeus).
Morphology
The genus Streptomyces includes aerobic, Gram-positive, multicellular, filamentous bacteria that produce well-developed vegetative hyphae (between 0.5-2.0 µm in diameter) with branches. They form a complex substrate mycelium that aids in scavenging organic compounds from their substrates.[15] Although the mycelia and the aerial hyphae that arise from them are amotile, mobility is achieved by dispersion of spores.[15] Spore surfaces may be hairy, rugose, smooth, spiny or warty.[16] In some species, aerial hyphae consist of long, straight filaments, which bear 50 or more spores at more or less regular intervals, arranged in whorls (verticils). Each branch of a verticil produces, at its apex, an umbel, which carries from two to several chains of spherical to ellipsoidal, smooth or rugose spores.[15] Some strains form short chains of spores on substrate hyphae. Sclerotia-, pycnidia-, sporangia-, and synnemata-like structures are produced by some strains.
Genomics
The complete genome of "S. coelicolor strain A3(2)" was published in 2002.[17] At the time, the "S. coelicolor" genome was thought to contain the largest number of genes of any bacterium.[17] The chromosome is 8,667,507 bp long with a GC-content of 72.1%, and is predicted to contain 7,825 protein-encoding genes.[17] In terms of taxonomy, "S. coelicolor A3(2)" belongs to the species S. violaceoruber, and is not a validly described separate species; "S. coelicolor A3(2)" is not to be mistaken for the actual S. coelicolor (Müller), although it is often referred to as S. coelicolor for convenience.[18] The transcriptome and translatome analyses of the strain A3(2) were published in 2016.[19]
The first complete genome sequence of S. avermitilis was completed in 2003.[20] Each of these genomes forms a chromosome with a linear structure, unlike most bacterial genomes, which exist in the form of circular chromosomes.[21] The genome sequence of S. scabiei, a member of the genus with the ability to cause potato scab disease, has been determined at the Wellcome Trust Sanger Institute. At 10.1 Mbp long and encoding 9,107 provisional genes, it is the largest known Streptomyces genome sequenced, probably due to the large pathogenicity island.[21][22]
The genomes of the various Streptomyces species demonstrate remarkable plasticity, via ancient single gene duplications, block duplications (mainly at the chromosomal arms) and horizontal gene transfer.[5][23] The size of their chromosome varies from 5.7-12.1 Mbps (average: 8.5 Mbps), the number of chromosomally encoded proteins varies from 4983-10,112 (average: 7130), whereas their high GC content varies from 68.8-74.7% (average: 71.7%).[5] The 95% soft-core proteome of the genus consists of approximately 2000-2400 proteins.[5] The pangenome is open.[24][25] In addition, significant genomic plasticity is observed even between strains of the same species, where the number of accessory proteins (at the species level) ranges from 250 to more than 3000.[5] Intriguingly, a correlation has been observed between the number of carbohydrate-active enzymes and secondary metabolite biosynthetic gene clusters (siderophores, e-Polylysin and type III lanthipeptides) that are related to competition among bacteria, in Streptomyces species.[5] Streptomycetes are major biomass degraders, mainly via their carbohydrate-active enzymes.[26] Thus, they also need to evolve an arsenal of siderophores and antimicrobial agents to suppress competition by other bacteria in these nutrient-rich environments that they create.[5] Several evolutionary analyses have revealed that the majority of evolutionarily stable genomic elements are localized mainly at the central region of the chromosome, whereas the evolutionarily unstable elements tend to localize at the chromosomal arms.[5][27][28][29][30] Thus, the chromosomal arms emerge as the part of the genome that is mainly responsible for rapid adaptation at both the species and strain level.[5]
Biotechnology
Biotechnology researchers have used Streptomyces species for heterologous expression of proteins. Traditionally, Escherichia coli was the species of choice to express eukaryotic genes, since it was well understood and easy to work with.[31][32] Expression of eukaryotic proteins in E. coli may be problematic. Sometimes, proteins do not fold properly, which may lead to insolubility, deposition in inclusion bodies, and loss of bioactivity of the product.[33] Though E. coli strains have secretion mechanisms, these are of low efficiency and result in secretion into the periplasmic space, whereas secretion by a Gram-positive bacterium such as a Streptomyces species results in secretion directly into the extracellular medium. In addition, Streptomyces species have more efficient secretion mechanisms than E.coli. The properties of the secretion system is an advantage for industrial production of heterologously expressed protein because it simplifies subsequent purification steps and may increase yield. These properties among others make Streptomyces spp. an attractive alternative to other bacteria such as E. coli and Bacillus subtilis.[33] In addition, the inherently high genomic instability suggests that the various Streptomycetes genomes may be amenable to extensive genome reduction for the construction of synthetic minimal genomes with industrial applications.[5]
Plant pathogenic bacteria
Several species belonging to this genus have been found to be pathogenic to plants:[12]
- S. scabiei
- S. acidiscabies
- S. europaeiscabiei
- S. luridiscabiei
- S. niveiscabiei
- S. puniciscabiei
- S. reticuliscabiei
- S. stelliscabiei
- S. turgidiscabies (scab disease in potatoes)
- S. ipomoeae (soft rot disease in sweet potatoes)
- S. brasiliscabiei (first species identified in Brazil)[34]
- S. hilarionis and S. hayashii (new species identified in Brazil)[35]
Medicine
Streptomyces is the largest antibiotic-producing genus, producing antibacterial, antifungal, and antiparasitic drugs, and also a wide range of other bioactive compounds, such as immunosuppressants.[36] Almost all of the bioactive compounds produced by Streptomyces are initiated during the time coinciding with the aerial hyphal formation from the substrate mycelium.[15]
Antifungals
Streptomycetes produce numerous antifungal compounds of medicinal importance, including nystatin (from S. noursei), amphotericin B (from S. nodosus),[37] and natamycin (from S. natalensis).
Antibacterials
Members of the genus Streptomyces are the source for numerous antibacterial pharmaceutical agents; among the most important of these are:
- Chloramphenicol (from S. venezuelae)[38]
- Daptomycin (from S. roseosporus)[39]
- Fosfomycin (from S. fradiae)[40]
- Lincomycin (from S. lincolnensis)[41]
- Neomycin (from S. fradiae)[42]
- Nourseothricin [citation needed]
- Puromycin (from S. alboniger)[43]
- Streptomycin (from S. griseus)[44]
- Tetracycline (from S. rimosus and S. aureofaciens)[45]
- Oleandomycin (from S. antibioticus)[46][47][48]
- Tunicamycin (from S. torulosus)[49]
- Mycangimycin (from Streptomyces sp. SPB74 and S. antibioticus)[50]
- Boromycin (from S. antibioticus)[51]
- Bambermycin (from S. bambergiensis and S. ghanaensis, the active compound being moenomycins A and C)[52]
- Vulgamycin[53]
Clavulanic acid (from S. clavuligerus) is a drug used in combination with some antibiotics (like amoxicillin) to block and/or weaken some bacterial-resistance mechanisms by irreversible beta-lactamase inhibition. Novel antiinfectives currently being developed include Guadinomine (from Streptomyces sp. K01-0509),[54] a compound that blocks the Type III secretion system of Gram-negative bacteria.
Antiparasitic drugs
S. avermitilis is responsible for the production of one of the most widely employed drugs against nematode and arthropod infestations, avermectin,[55] and thus its derivatives including ivermectin.
Other

Less commonly, streptomycetes produce compounds used in other medical treatments: migrastatin (from S. platensis) and bleomycin (from S. verticillus) are antineoplastic (anticancer) drugs; boromycin (from S. antibioticus) exhibits antiviral activity against the HIV-1 strain of HIV, as well as antibacterial activity. Staurosporine (from S. staurosporeus) also has a range of activities from antifungal to antineoplastic (via the inhibition of protein kinases).
S. hygroscopicus and S. viridochromogenes produce the natural herbicide bialaphos.
Saptomycins are chemical compounds isolated from Streptomyces.[56]
Symbiosis
Sirex wasps cannot perform all of their own cellulolytic functions and so some Streptomyces do so in symbiosis with the wasps.[57] Book et al. have investigated several of these symbioses.[57] Book et al., 2014 and Book et al., 2016 identify several lytic isolates.[57] The 2016 study isolates Streptomyces sp. Amel2xE9 and Streptomyces sp. LamerLS-31b and finds that they are equal in activity to the previously identified Streptomyces sp. SirexAA-E.[57]
See also
- Antimycin A – Chemical compound produced by Streptomyces used as a piscicide
- Geosmin
- Streptomyces isolates
- List of bacterial orders
- List of bacteria genera
References
- ↑ "Streptomyces". List of Prokaryotic names with Standing in Nomenclature (LPSN). https://lpsn.dsmz.de/genus/streptomyces.
- ↑ Van der Meij, A., Willemse, J., Schneijderberg, M.A., Geurts, R., Raaijmakers, J.M. and van Wezel, G.P. (2018) "Inter-and intracellular colonization of Arabidopsis roots by endophytic actinobacteria and the impact of plant hormones on their antimicrobial activity". Antonie van Leeuwenhoek, 111(5): 679–690. doi:10.1007/s10482-018-1014-z
- ↑ "The Family Streptomycetaceae, Part I: Taxonomy". The Prokaryotes. 2006. pp. 538–604. doi:10.1007/0-387-30743-5_22. ISBN 978-0-387-25493-7. https://books.google.com/books?id=swciHNNWZDEC&pg=PA538.
- ↑ "Genus Streptomyces". List of Prokaryotic names with Standing in Nomenclature. 2008. http://www.bacterio.cict.fr/s/streptomycesa.html.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 Nikolaidis, Marios; Hesketh, Andrew; Frangou, Nikoletta; Mossialos, Dimitris; Van de Peer, Yves; Oliver, Stephen G.; Amoutzias, Grigorios D. (June 2023). "A panoramic view of the genomic landscape of the genus Streptomyces". Microbial Genomics 9 (6). doi:10.1099/mgen.0.001028. ISSN 2057-5858. PMID 37266990.
- ↑ 6.0 6.1 "Genus: Streptomyces" (in en). https://www.bacterio.net/genus/streptomyces.
- ↑ 7.0 7.1 Brock Biology of Microorganisms (11th ed.). Prentice Hall. 2005. ISBN 978-0-13-144329-7.[page needed]
- ↑ John Wiley & Sons, Ltd, ed (2001-05-30) (in en). eLS (1 ed.). Wiley. doi:10.1002/9780470015902.a0020392.pub2. ISBN 978-0-470-01617-6. https://onlinelibrary.wiley.com/doi/book/10.1002/047001590X.
- ↑ Practical Streptomyces Genetics (2nd ed.). Norwich, England: John Innes Foundation. 2000. ISBN 978-0-7084-0623-6.[page needed]
- ↑ "Understanding and manipulating antibiotic production in actinomycetes". Biochemical Society Transactions 41 (6): 1355–64. December 2013. doi:10.1042/BST20130214. PMID 24256223.
- ↑ "The taxonomy of Streptomyces and related genera". International Journal of Systematic and Evolutionary Microbiology 51 (Pt 3): 797–814. May 2001. doi:10.1099/00207713-51-3-797. PMID 11411701.
- ↑ 12.0 12.1 "Multilocus sequence analysis of phytopathogenic species of the genus Streptomyces". International Journal of Systematic and Evolutionary Microbiology 61 (Pt 10): 2525–2531. October 2011. doi:10.1099/ijs.0.028514-0. PMID 21112986.
- ↑ "A proposal to revive the genus Kitasatospora (Omura, Takahashi, Iwai, and Tanaka 1982)". International Journal of Systematic Bacteriology 47 (4): 1048–54. October 1997. doi:10.1099/00207713-47-4-1048. PMID 9336904.
- ↑ "Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici (1943)AL) emend. Rainey et al. 1997". Antonie van Leeuwenhoek 83 (2): 107–16. 2003. doi:10.1023/A:1023397724023. PMID 12785304.
- ↑ 15.0 15.1 15.2 15.3 "Morphological and physiological differentiation in Streptomyces". Microbial development. 16. 1984. pp. 89–115. doi:10.1101/0.89-115. ISBN 978-0-87969-172-1. http://cshmonographs.org/index.php/monographs/article/view/4367. Retrieved 2012-01-19.
- ↑ "Classification of Streptomyces spore surfaces into five groups". Applied Microbiology 21 (3): 527–33. March 1971. doi:10.1128/AEM.21.3.527-533.1971. PMID 4928607.
- ↑ 17.0 17.1 17.2 "Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2)". Nature 417 (6885): 141–7. May 2002. doi:10.1038/417141a. PMID 12000953. Bibcode: 2002Natur.417..141B.
- ↑ "The complex extracellular biology of Streptomyces". FEMS Microbiology Reviews 34 (2): 171–98. March 2010. doi:10.1111/j.1574-6976.2009.00206.x. PMID 20088961.
- ↑ "The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2)". Nature Communications 7 (1): 11605. June 2016. doi:10.1038/ncomms11605. PMID 27251447. Bibcode: 2016NatCo...711605J.
- ↑ "Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis". Nature Biotechnology 21 (5): 526–31. May 2003. doi:10.1038/nbt820. PMID 12692562.
- ↑ 21.0 21.1 Streptomyces: Molecular Biology and Biotechnology. Horizon Scientific Press. 1 January 2011. p. 5. ISBN 978-1-904455-77-6. https://books.google.com/books?id=3z9_QwFumi8C. Retrieved 16 January 2012.
- ↑ "Streptomyces scabies". Sanger Institute. http://www.sanger.ac.uk/Projects/S_scabies.
- ↑ McDonald, Bradon R.; Currie, Cameron R. (2017-06-06). "Lateral Gene Transfer Dynamics in the Ancient Bacterial Genus Streptomyces". mBio 8 (3): e00644–17. doi:10.1128/mBio.00644-17. ISSN 2150-7511. PMID 28588130.
- ↑ Caicedo-Montoya, Carlos; Manzo-Ruiz, Monserrat; Ríos-Estepa, Rigoberto (2021). "Pan-Genome of the Genus Streptomyces and Prioritization of Biosynthetic Gene Clusters With Potential to Produce Antibiotic Compounds". Frontiers in Microbiology 12: 677558. doi:10.3389/fmicb.2021.677558. ISSN 1664-302X. PMID 34659136.
- ↑ Otani, Hiroshi; Udwary, Daniel W.; Mouncey, Nigel J. (2022-11-07). "Comparative and pangenomic analysis of the genus Streptomyces". Scientific Reports 12 (1): 18909. doi:10.1038/s41598-022-21731-1. ISSN 2045-2322. PMID 36344558. Bibcode: 2022NatSR..1218909O.
- ↑ Chater, Keith F.; Biró, Sandor; Lee, Kye Joon; Palmer, Tracy; Schrempf, Hildgund (March 2010). "The complex extracellular biology of Streptomyces". FEMS Microbiology Reviews 34 (2): 171–198. doi:10.1111/j.1574-6976.2009.00206.x. ISSN 1574-6976. PMID 20088961.
- ↑ Lorenzi, Jean-Noël; Lespinet, Olivier; Leblond, Pierre; Thibessard, Annabelle (September 2019). "Subtelomeres are fast-evolving regions of the Streptomyces linear chromosome". Microbial Genomics 7 (6): 000525. doi:10.1099/mgen.0.000525. ISSN 2057-5858. PMID 33749576.
- ↑ Tidjani, Abdoul-Razak; Lorenzi, Jean-Noël; Toussaint, Maxime; van Dijk, Erwin; Naquin, Delphine; Lespinet, Olivier; Bontemps, Cyril; Leblond, Pierre (2019-09-03). "Massive Gene Flux Drives Genome Diversity between Sympatric Streptomyces Conspecifics". mBio 10 (5): e01533–19. doi:10.1128/mBio.01533-19. ISSN 2150-7511. PMID 31481382.
- ↑ Volff, J. N.; Altenbuchner, J. (January 1998). "Genetic instability of the Streptomyces chromosome". Molecular Microbiology 27 (2): 239–246. doi:10.1046/j.1365-2958.1998.00652.x. ISSN 0950-382X. PMID 9484880. https://pubmed.ncbi.nlm.nih.gov/9484880.
- ↑ Chen, Carton W.; Huang, Chih-Hung; Lee, Hsuan-Hsuan; Tsai, Hsiu-Hui; Kirby, Ralph (October 2002). "Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes". Trends in Genetics 18 (10): 522–529. doi:10.1016/s0168-9525(02)02752-x. ISSN 0168-9525. PMID 12350342. https://pubmed.ncbi.nlm.nih.gov/12350342.
- ↑ "Streptomyces: a host for heterologous gene expression". Current Opinion in Biotechnology 2 (5): 674–81. October 1991. doi:10.1016/0958-1669(91)90033-2. PMID 1367716.
- ↑ "Improved production of heterologous protein from Streptomyces lividans". Applied Microbiology and Biotechnology 33 (4): 395–400. July 1990. doi:10.1007/BF00176653. PMID 1369282.
- ↑ 33.0 33.1 "Heterologous biopharmaceutical protein expression in Streptomyces". Trends in Biotechnology 15 (8): 315–20. August 1997. doi:10.1016/S0167-7799(97)01062-7. PMID 9263479.
- ↑ Corrêa, Daniele Bussioli Alves; do Amaral, Danilo Trabuco; da Silva, Márcio José; Destéfano, Suzete Aparecida Lanza (July 2021). "Streptomyces brasiliscabiei, a new species causing potato scab in south Brazil". Antonie van Leeuwenhoek 114 (7): 913–931. doi:10.1007/s10482-021-01566-y. PMID 33881637.
- ↑ Vitor, Lucas; Amaral, Danilo Trabuco; Corrêa, Daniele Bussioli Alves; Ferreira-Tonin, Mariana; Lucon, Emanuel Torres; Appy, Mariana Pereira; Tomaseto, Alex Augusto; Destéfano, Suzete Aparecida Lanza (15 June 2023). "Streptomyces hilarionis sp. nov. and Streptomyces hayashii sp. nov., two new strains associated with potato scab in Brazil". International Journal of Systematic and Evolutionary Microbiology 73 (6). doi:10.1099/ijsem.0.005916. PMID 37319004.
- ↑ "How many antibiotics are produced by the genus Streptomyces?". Archives of Microbiology 176 (5): 386–90. November 2001. doi:10.1007/s002030100345. PMID 11702082.
- ↑ "Antibiotics produced by Streptomyces". The Brazilian Journal of Infectious Diseases 16 (5): 466–71. 2012. doi:10.1016/j.bjid.2012.08.014. PMID 22975171.
- ↑ "A plasmid involved in chloramphenicol production in Streptomyces venezuelae: evidence from genetic mapping". Journal of General Microbiology 90 (2): 336–46. October 1975. doi:10.1099/00221287-90-2-336. PMID 1194895.
- ↑ "Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry". Microbiology 151 (Pt 5): 1507–1523. May 2005. doi:10.1099/mic.0.27757-0. PMID 15870461.
- ↑ "Heterologous production of fosfomycin and identification of the minimal biosynthetic gene cluster". Chemistry & Biology 13 (11): 1171–82. November 2006. doi:10.1016/j.chembiol.2006.09.007. PMID 17113999.
- ↑ "Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11". Molecular Microbiology 16 (6): 1137–56. June 1995. doi:10.1111/j.1365-2958.1995.tb02338.x. PMID 8577249.
- ↑ "The production of neomycin by Streptomyces fradiae in synthetic media". Applied Microbiology 1 (2): 103–6. March 1953. doi:10.1128/AEM.1.2.103-106.1953. PMID 13031516.
- ↑ "Biosynthesis of puromycin in Streptomyces alboniger: regulation and properties of O-demethylpuromycin O-methyltransferase". Antimicrobial Agents and Chemotherapy 8 (6): 721–32. December 1975. doi:10.1128/AAC.8.6.721. PMID 1211926.
- ↑ "Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity". Nucleic Acids Research 15 (19): 8041–56. October 1987. doi:10.1093/nar/15.19.8041. PMID 3118332.
- ↑ Tetracyclines in biology, chemistry and medicine. Birkhäuser. 2001. pp. 8–. ISBN 978-3-7643-6282-9. https://books.google.com/books?id=kHNW4tFhZD4C&pg=PA8. Retrieved 17 January 2012.
- ↑ "What are Streptomycetes?". http://home.hiroshima-u.ac.jp/mbiotech/hosenkin_lab/Strepto-E.html.
- ↑ "Characterisation of a Streptomyces antibioticus gene encoding a type I polyketide synthase which has an unusual coding sequence". Molecular & General Genetics 242 (3): 358–62. February 1994. doi:10.1007/BF00280426. PMID 8107683.
- ↑ "Finto: MeSH: Streptomyces antibioticus". http://finto.fi/mesh/en/page/D013303.
- ↑ "Biochemical studies on antibiotic production from Streptomyces sp.: Taxonomy, fermentation, isolation and biological properties". Journal of Saudi Chemical Society 19 (1): 12–22. January 2015. doi:10.1016/j.jscs.2011.12.011.
- ↑ "Mycangimycin, a polyene peroxide from a mutualist Streptomyces sp". Organic Letters 11 (3): 633–6. February 2009. doi:10.1021/ol802709x. PMID 19125624.
- ↑ "Biosynthesis of boromycin". The Journal of Organic Chemistry 46 (13): 2661–2665. June 1981. doi:10.1021/jo00326a010.
- ↑ "CID=53385491". PubChem Compound Database. National Center for Biotechnology Information.. https://pubchem.ncbi.nlm.nih.gov/compound/53385491.
- ↑ Babczinski, Peter; Dorgerloh, Michael; Löbberding, Antonius; Santel, Hans-Joachim; Schmidt, Robert R.; Schmitt, Peter; Wünsche, Christian (1991). "Herbicidal activity and mode of action of vulgamycin" (in en). Pesticide Science 33 (4): 439–446. doi:10.1002/ps.2780330406. https://onlinelibrary.wiley.com/doi/10.1002/ps.2780330406.
- ↑ "Molecular insights into the biosynthesis of guadinomine: a type III secretion system inhibitor". Journal of the American Chemical Society 134 (42): 17797–806. October 2012. doi:10.1021/ja308622d. PMID 23030602.
- ↑ Martín, Juan F; Rodríguez-García, Antonio; Liras, Paloma (2017-03-15). "The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: comparison in Streptomyces coelicolor and Streptomyces avermitilis". The Journal of Antibiotics (Japan Antibiotics Research Association (Nature Portfolio)) 70 (5): 534–541. doi:10.1038/ja.2017.19. ISSN 0021-8820. PMID 28293039.
- ↑ Abe, N.; Nakakita, Y.; Nakamura, T.; Enoki, N.; Uchida, H.; Munekata, M. (1993). "Novel antitumor antibiotics, saptomycins. I. Taxonomy of the producing organism, fermentation, HPLC analysis and biological activities". The Journal of Antibiotics 46 (10): 1530–5. doi:10.7164/antibiotics.46.1530. PMID 8244880.
- ↑ 57.0 57.1 57.2 57.3 Li, Hongjie; Young, Soleil E.; Poulsen, Michael; Currie, Cameron R. (2021-01-07). "Symbiont-Mediated Digestion of Plant Biomass in Fungus-Farming Insects". Annual Review of Entomology (Annual Reviews) 66 (1): 297–316. doi:10.1146/annurev-ento-040920-061140. ISSN 0066-4170. PMID 32926791.
Further reading
- Genetics and Product Formation in Streptomyces. Kluwer Academic. 1991. ISBN 978-0-306-43885-1.
- Bacteria: Antibiotic-producing Streptomyces. Academic Press. 1986. ISBN 978-0-12-307209-2.
- Streptomyces in Nature and Medicine: The Antibiotic Makers. Oxford University Press. 2007. ISBN 978-0-19-515066-7.
- Streptomyces: Molecular Biology and Biotechnology. Caister Academic Press. 2011. ISBN 978-1-904455-77-6.
External links
- "Current research on Streptomyces coelicolor". Norwich Research Park. 3 January 2018. http://www.micron.ac.uk/organisms/sco.html.
- "Some current Streptomyces Research & Methods / Protocols / Resources". www.openwetware.org. http://www.openwetware.org/wiki/Streptomyces.
- "S. avermitilis genome homepage". Kitasato Institute for Life Sciences. http://avermitilis.ls.kitasato-u.ac.jp/.
- "S. coelicolor A3(2) genome homepage". Sanger Institute. http://www.sanger.ac.uk/Projects/S_coelicolor/.
- "Streptomyces.org.uk homepage". John Innes Centre. http://streptomyces.org.uk/.
- "StrepDB - the Streptomyces genomes annotation browser". http://strepdb.streptomyces.org.uk/.
- "Streptomyces Genome Projects". Genomes OnLine Database. http://www.genomesonline.org/search.cgi?colcol=all&goldstamp=ALL&gen_type=ALL&org_name1=genus&gensp=Streptomyces&org_domain=ALL&org_status=ALL&size2=ALL&org_size=Kb&gen_gc=ALL&phylogeny2=ALL&gen_institution=ALL&gen_funding=ALL&gen_data=ALL&cont=ALL&gen_country=ALL&gen_pheno=ALL&gen_eco=ALL&gen_disease=ALL&gen_relevance=ALL&gen_avail=ALL&selection=submit+search.
Wikidata ☰ Q1144013 entry
 |
