Medicine:WRAP53
 Generic protein structure example |
WRAP53 (also known as WD40-encoding RNA antisense to p53) is a gene implicated in cancer development. The name was coined in 2009 to describe the dual role of this gene, encoding both an antisense RNA that regulates the p53 tumor suppressor and a protein involved in DNA repair, telomere elongation and maintenance of nuclear organelles Cajal bodies (Figure 1).[1][2][3][4]
Gene
The WRAP53 gene is localized on chromosome 17p13.1 and contains 13 exons, including three alternative starting exons (1α, 1β and 1γ) generating at least three gene products. The WRAP53 gene partially overlaps the p53 tumor suppressor gene in a head-to-head orientation.
Function

Transcripts of WRAP53 that overlap the first exon of p53 (referred to as WRAP53α transcripts) regulate the levels of p53 mRNA and protein (Figure 1).[1][5] WRAP53γ transcripts overlap the first intron of p53 and are antisense to the previously identified transcript Hp53int1 localized within this intron. However, the function of WRAP53γ remains elusive.
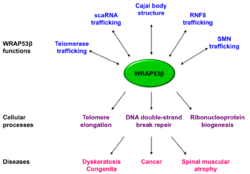
The WRAP53 gene also encodes a protein termed WRAP53β (alias WRAP53 or WDR79 or TCAB1), which belongs to the WD40 protein family (Figure 1). WRAP53β facilitates protein-protein and protein-RNA interactions and directs factors to nuclear organelles Cajal bodies, to telomeres and to DNA double-strand breaks.[3][4][6][7] Factors localized to Cajal bodies with the help of WRAP53β includes the SMN protein,[6] small Cajal body-specific scaRNAs[4] and the enzyme telomerase.[3] WRAP53β also targets the ubiquitin ligase RNF8 to DNA double-strand breaks[7](Figure 2).
In addition to localizing factors to correct cellular sites, WRAP53β maintain structural integrity of Cajal bodies and without WRAP53β these organelles collapse and cannot re-form [6] (Figure 2).
Domains

The WRAP53β protein is highly evolutionary conserved, with homologs (confined to its WD40 repeats) in vertebrates, invertebrates, plants and yeast.[1][8][9] WRAP53β consists of a proline-rich N-terminus, a central WD40 domain and a glycine-rich C-terminus (Figure 3). The WD40 domain of WRAP53β serves as a scaffold for multiple interactions between a wide variety of molecules.
Clinical significance
Dyskeratosis congenita
Germline mutations in WRAP53β result in disorder known as dyskeratosis congenita, characterized by bone marrow failure, premature ageing, predisposition for cancer and a triad of mucocutaneous features including oral leukoplakia, abnormal skin pigmentation and nail dystrophy.[9] Mutations in WRAP53β are inherited in an autosomal recessive fashion, reside in highly conserved regions of its WD40 domain and result in a more severe form of this disease.[10][11]
These mutations reduce the nuclear level of WRAP53β, impair its trafficking of telomerase to telomeres, and subsequently lead to progressive shortening of telomeres in these patients [9] The chaperonin CCT/TRiC is crucial for proper folding of WRAP53β and this folding is impaired in dyskeratosis congenita[12]
Spinal muscular atrophy
Defective WRAP53β-mediated trafficking of SMN is observed in patients afflicted by the most severe form of spinal muscular atrophy (type I or Werdnig-Hoffmann disease),[13][14] a neurodegenerative disorder characterized by progressive degeneration of spinal cord anterior horn α-motor neurons and the leading genetic cause of infant mortality with an incidence of approximately 1:6000 live births.[15] Mutations in the SMN1 gene are the underlying cause to spinal muscular atrophy (SMA).
Cancer
WRAP53β is overexpressed in a variety of cancer cell lines of different origins and such overexpression promotes carcinogenic transformation indicating that this protein possesses oncogenic properties.[16] WRAP53β is overexpressed in primary nasopharyngeal carcinoma,[17] esophageal squamous cell carcinoma[18] and rectal cancer.[19] Moreover, knockdown of WRAP53β in cancer cells reduced the size of the tumors formed when these are grafted into mice [17] and triggers mitochondrial-dependent apoptosis in cancer cells.[16]
In contrary, inactivating mutations in both alleles of WRAP53β causes dyskeratosis congenita, indicating that this protein acts as tumor suppressor, rather than an oncogene. Loss of nuclear WRAP53β is also correlated with shortened survival and resistance to radiotherapy in patients with head and neck cancer.[20] With its complex roles in a number of cellular processes, WRAP53β may act as a tumor suppressor under certain conditions and as an oncogene in under others.
Single nucleotide polymorphisms (SNPs) in the WRAP53 gene have been linked to an increased risk for breast and ovarian cancer.[21][22][23] One of these SNPs also associate with defective DNA repair and hematotoxicity in workers exposed to benzene.[24]
Discovery
The WRAP53 gene was first identified by Marianne Farnebo (maiden name Hammarsund) at Karolinska Institutet in 2006. In 2009 the WRAP53β (alias TCAB1, WDR79 and WRAP53) protein was described by Steven Artandi (Stanford University), Joan Steitz (Yale University) and Marianne Farnebo (Karolinska Institutet) independently.
References
- ↑ 1.0 1.1 1.2 "Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage". Molecular Cell 33 (4): 462–71. February 2009. doi:10.1016/j.molcel.2009.01.028. PMID 19250907.
- ↑ "Wrap53, a novel regulator of p53". Cell Cycle (Georgetown, Tex.) 8 (15): 2343–6. August 2009. doi:10.4161/cc.8.15.9223. PMID 19571673.
- ↑ 3.0 3.1 3.2 "A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis". Science (New York, N.Y.) 323 (5914): 644–8. January 2009. doi:10.1126/science.1165357. PMID 19179534. Bibcode: 2009Sci...323..644V.
- ↑ 4.0 4.1 4.2 "A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles". Molecular Cell 34 (1): 47–57. April 2009. doi:10.1016/j.molcel.2009.02.020. PMID 19285445.
- ↑ "CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53". Genes & Development 28 (7): 723–34. April 2014. doi:10.1101/gad.236869.113. PMID 24696455.
- ↑ 6.0 6.1 6.2 "WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies". PLOS Biology 8 (11): e1000521. November 2010. doi:10.1371/journal.pbio.1000521. PMID 21072240.
- ↑ 7.0 7.1 "The scaffold protein WRAP53β orchestrates the ubiquitin response critical for DNA double-strand break repair". Genes & Development 28 (24): 2726–38. December 2014. doi:10.1101/gad.246546.114. PMID 25512560.
- ↑ "A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles". Molecular Cell 34 (1): 47–57. April 2009. doi:10.1016/j.molcel.2009.02.020. PMID 19285445.
- ↑ 9.0 9.1 9.2 "Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita". Genes & Development 25 (1): 11–6. January 2011. doi:10.1101/gad.2006411. PMID 21205863.
- ↑ "Dyskeratosis congenita". Hematology. American Society of Hematology. Education Program 2011: 480–6. 2011. doi:10.1182/asheducation-2011.1.480. PMID 22160078.
- ↑ "Updates on the biology and management of dyskeratosis congenita and related telomere biology disorders". Expert Review of Hematology 6 (3): 327–37. June 2013. doi:10.1586/ehm.13.23. PMID 23782086. https://zenodo.org/record/1235762.
- ↑ "Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1". Cell 159 (6): 1389–403. December 2014. doi:10.1016/j.cell.2014.10.059. PMID 25467444.
- ↑ "Correlation between severity and SMN protein level in spinal muscular atrophy". Nature Genetics 16 (3): 265–9. July 1997. doi:10.1038/ng0797-265. PMID 9207792.
- ↑ "Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy". Histochemistry and Cell Biology 137 (5): 657–67. May 2012. doi:10.1007/s00418-012-0921-8. PMID 22302308.
- ↑ "SMN in spinal muscular atrophy and snRNP biogenesis". Wiley Interdisciplinary Reviews. RNA 2 (4): 546–64. 2011. doi:10.1002/wrna.76. PMID 21957043.
- ↑ 16.0 16.1 "WRAP53 promotes cancer cell survival and is a potential target for cancer therapy". Cell Death & Disease 2 (1): e114. January 2011. doi:10.1038/cddis.2010.90. PMID 21368886.
- ↑ 17.0 17.1 "TCAB1: a potential target for diagnosis and therapy of head and neck carcinomas". Molecular Cancer 13: 180. July 2014. doi:10.1186/1476-4598-13-180. PMID 25070141.
- ↑ "Overexpression of WRAP53 is associated with development and progression of esophageal squamous cell carcinoma". PLOS ONE 9 (3): e91670. 2014. doi:10.1371/journal.pone.0091670. PMID 24626331. Bibcode: 2014PLoSO...991670R.
- ↑ "WRAP53 is an independent prognostic factor in rectal cancer- a study of Swedish clinical trial of preoperative radiotherapy in rectal cancer patients". BMC Cancer 12: 294. July 2012. doi:10.1186/1471-2407-12-294. PMID 22805008.
- ↑ "Nuclear expression of WRAP53β is associated with a positive response to radiotherapy and improved overall survival in patients with head and neck squamous cell carcinoma". Oral Oncology 51 (1): 24–30. January 2015. doi:10.1016/j.oraloncology.2014.10.003. PMID 25456005.
- ↑ "Common genetic variation in TP53 and its flanking genes, WDR79 and ATP1B2, and susceptibility to breast cancer". International Journal of Cancer 121 (11): 2532–8. December 2007. doi:10.1002/ijc.22985. PMID 17683073.
- ↑ "Association of common WRAP 53 variant with ovarian cancer risk in the Polish population". Molecular Biology Reports 40 (3): 2145–7. March 2013. doi:10.1007/s11033-012-2273-9. PMID 23192612.
- ↑ "Single nucleotide polymorphisms in the TP53 region and susceptibility to invasive epithelial ovarian cancer". Cancer Research 69 (6): 2349–57. March 2009. doi:10.1158/0008-5472.CAN-08-2902. PMID 19276375.
- ↑ "Large-scale evaluation of candidate genes identifies associations between DNA repair and genomic maintenance and development of benzene hematotoxicity". Carcinogenesis 30 (1): 50–8. January 2009. doi:10.1093/carcin/bgn249. PMID 18978339.
 |
