Software:Disordered Structure Refinement
The Disordered Structure Refinement[1][2] program (DSR), written by Daniel Kratzert, is designed to simplify the modeling of molecular disorder in crystal structures using SHELXL[3][4][5] by George M. Sheldrick. It has a database of approximately 120 standard solvent molecules and molecular moieties. These can be inserted into the crystal structure with little effort, while at the same time chemically meaningful binding and angular restraints[6] are set. DSR was developed because the previous description of disorder in crystal structures with SHELXL was very lengthy and error-prone. Instead of editing large text files manually and defining restraints manually, this process is automated with DSR.
Application
DSR can be started in a command line. The call has the basic form:
dsr [option] (SHELXL file)
DSR is controlled with a special command in the respective SHELXL file. This has the following syntax:
REM DSR PUT/REPLACE "fragment" WITH (atoms) ON (atoms oder q-peaks) PART 1 OCC -21 = RESI DFIX
The DSR command must always start with REM so that SHELXL does not recognize this line as own command. Behind WITH and ON stands which atom of the molecule fragment from the database to which atom or q-peak is to be positioned in the crystal structure.
By running
dsr -r file.res
the fragment fit is performed and the restraints transferred.
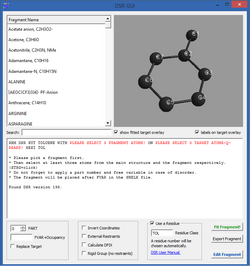
Graphical User Interface
Since 2016 ShelXle has a graphical interface to DSR. Most commands of the command line version can be executed there.
In order to transfer a fragment into a structure, three atoms / q-peaks have to be selected in ShelXle and in the DSR GUI each to specify the position of the fragment. The 3D view of the fragment then shows a preview of the subsequent fragment fit.
Programming
DSR is only programmed in Python. Therefore, it runs in any Python-supported operating system.
It is under the free Beerware license and can be downloaded free of charge and changed as desired.
External links
- The source code on Github Github.
- Official Web site.
References
- ↑ Kratzert, Daniel; Krossing, Ingo (3 May 2018). "Recent improvements in DSR". Journal of Applied Crystallography 51 (3): 928–934. doi:10.1107/S1600576718004508. ISSN 1600-5767. http://journals.iucr.org/j/issues/2018/03/00/to5174/index.html. Retrieved 17 August 2018.
- ↑ Daniel Kratzert, Julian J. Holstein, Ingo Krossing (2015-06-01), "DSR: enhanced modelling and refinement of disordered structures withSHELXL" (in German), Journal of Applied Crystallography 48 (3): 933–938, doi:10.1107/s1600576715005580, ISSN 1600-5767, PMID 26089767, ISSN 1600-5767, http://scripts.iucr.org/cgi-bin/paper?S1600576715005580. Retrieved 2017-02-07
- ↑ "The SHELX homepage" (in German). http://shelx.uni-goettingen.de/. Retrieved 2017-02-07.
- ↑ George M. Sheldrick (2008-01-01), "A short history ofSHELX" (in German), Acta Crystallographica Section a Foundations of Crystallography 64 (1): 112–122, doi:10.1107/s0108767307043930, ISSN 0108-7673, PMID 18156677, Bibcode: 2008AcCrA..64..112S, ISSN 0108-7673
- ↑ George M. Sheldrick (2015-01-01), "Crystal structure refinement withSHELXL" (in German), Acta Crystallographica Section C Structural Chemistry 71 (1): 3–8, doi:10.1107/s2053229614024218, ISSN 2053-2296, PMID 25567568, ISSN 2053-2296
- ↑ Gerard J. Kleywegt (2007-01-01), "Crystallographic refinement of ligand complexes" (in German), Acta Crystallographica Section D Biological Crystallography 63 (1): 94–100, doi:10.1107/s0907444906022657, ISSN 0907-4449, PMID 17164531, ISSN 0907-4449