Biology:Parabacteroides-1 RNA motif
From HandWiki
| Parabacteroides-1 | |
|---|---|
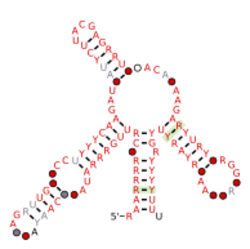 Consensus secondary structure and sequence conservation of Parabacteroides-1 RNA | |
| Identifiers | |
| Symbol | Parabacteroides-1 |
| Rfam | RF03088 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0001263 |
| PDB structures | PDBe |
The Parabacteroides-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] Energetically stable tetraloops often occur in this motif. Parabacteroides-1 motif RNAs are found in Parabacteroides.
Parabacteroides-1 RNAs likely function in trans as small RNAs, and are immediately followed by a predicted Rho-independent transcription terminator hairpin. Based on the protein-coding genes that are nearby to Parabacteroides-1 RNAs, it is likely that at least some of these RNAs are located in prophage region.
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Research 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
 |

