Medicine:Tetramer assay
| Tetramer assay | |
|---|---|
| Medical diagnostics | |
| Synonyms | Tetramer stain |
| Purpose | uses tetrameric proteins to detect and quantify T cells |
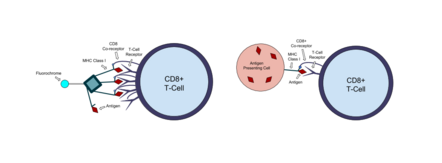
A tetramer assay (also known as a tetramer stain) is a procedure that uses tetrameric proteins to detect and quantify T cells that are specific for a given antigen within a blood sample.[1] The tetramers used in the assay are made up of four major histocompatibility complex (MHC) molecules, which are found on the surface of most cells in the body.[2] MHC molecules present peptides to T-cells as a way to communicate the presence of viruses, bacteria, cancerous mutations, or other antigens in a cell. If a T-cell's receptor matches the peptide being presented by an MHC molecule, an immune response is triggered.[3] Thus, MHC tetramers that are bioengineered to present a specific peptide can be used to find T-cells with receptors that match that peptide. The tetramers are labeled with a fluorophore, allowing tetramer-bound T-cells to be analyzed with flow cytometry.[4] Quantification and sorting of T-cells by flow cytometry enables researchers to investigate immune response to viral infection and vaccine administration as well as functionality of antigen-specific T-cells.[5] Generally, if a person's immune system has encountered a pathogen, the individual will possess T cells with specificity toward some peptide on that pathogen. Hence, if a tetramer stain specific for a pathogenic peptide results in a positive signal, this may indicate that the person's immune system has encountered and built a response to that pathogen.[citation needed]
History
This methodology was first published in 1996 by a lab at Stanford University.[6] Previous attempts to quantify antigen-specific T-cells involved the less accurate limiting dilution assay, which estimates numbers of T-cells at 50-500 times below their actual levels.[7][8] Stains using soluble MHC monomers were also unsuccessful due to the low binding affinity of T-cell receptors and MHC-peptide monomers. MHC tetramers can bind to more than one receptor on the target T-cell, resulting in an increased total binding strength and lower dissociation rates.[5]
Uses
CD8+ T-cells
Tetramer stains usually analyze cytotoxic T lymphocyte (CTL) populations.[9] CTLs are also called CD8+ T-cells, because they have CD8 co-receptors that bind to MHC class I molecules. Most cells in the body express MHC class I molecules, which are responsible for processing intracellular antigens and presenting at the cell's surface. If the peptides being presented by MHC class I molecules are foreign—for example, derived from viral proteins instead of the cell's own proteins—the CTL with a receptor that matches the peptide will destroy the cell.[2][3] Tetramer stains allow for the visualization, quantification, and sorting of these cells by flow cytometry, which is extremely useful in immunology. T-cell populations can be tracked over the duration of a virus or after the application of a vaccine. Tetramer stains can also be paired with functional assays like ELIspot, which detects the number of cytokine secreting cells in a sample.[9]
MHC Class I Tetramer Construction
MHC tetramer molecules developed in a lab can mimic the antigen presenting complex on cells and bind to T-cells that recognize the antigen. Class I MHC molecules are made up of a polymorphic heavy α-chain associated with an invariant light chain beta-2 microglobulin (β2m). Escherichia coli are used to synthesize the light chain and a shortened version of the heavy chain that includes the biotin 15 amino acid recognition tag. These MHC chains are biotinylated with the enzyme BirA and refolded with the antigenic peptide of interest. Biotin is a small molecule that forms a strong bond with another protein called streptavidin. Fluorophore tagged streptavidin is added to the bioengineered MHC monomers, and the biotin-streptavidin interaction causes four MHC monomers to bind to the streptavidin and create a tetramer. When the tetramers are mixed with a blood sample, they will bind to T-cells expressing the appropriate antigen specific receptor. Any MHC tetramers that are not bound are washed out of the sample before it is analyzed with flow cytometry.[9]
Recent advancements within recombinant MHC molecules have democratised peptide MHC complex formulation and subsequent multimerisation. Highly active formulations of a broad range of MHC class I molecules[10] now allows non-experts users to make their own custom peptide-MHC complexes from day-to-day in any lab without special equipment.[citation needed]
CD4+ T-cells
Tetramers that bind to helper T-cells have also been developed.[9] Helper T-cells or CD4+ T-cells express CD4 co-receptors. They bind to class II MHC molecules, which are only expressed in professional antigen-presenting cells like dendritic cells or macrophages. Class II MHC molecules present extracellular antigens, allowing helper T-cells to detect bacteria,[11] fungi, and parasites.[2] Class II MHC tetramer use is becoming more common, but the tetramers are more difficult to create than class I tetramers and the bond between helper T-cells and MHC molecules is even weaker.[9][12]
Natural Killer T-cells
Natural killer T-cells (NKT cells) can also be visualized with tetramer technology. NKT cells bind to proteins that present lipid or glycolipid antigens.[13] The antigen presenting complex that NKT cells bind to involves CD1 proteins, so tetramers made of CD1 can be used to stain for NKT cells.[9]
Examples
An early application of tetramer technology focused on the cell-mediated immune response to HIV infection. MHC tetramers were developed to present HIV antigens and used to find the percentage of CTLs specific to those HIV antigens in blood samples of infected patients. This was compared to results of cytotoxic assays and plasma RNA viral load to characterize the function of CTLs in HIV infection. The CTLs that bound to tetramers were sorted into ELIspot wells for analysis of cytokine secretion.[14]
Another study utilized MHC tetramer complexes to investigate the effectiveness of an influenza vaccine delivery method. Mice were given subcutaneous and intranasal vaccinations for influenza, and tetramer stains coupled with flow cytometry were used to quantify the CTLs specific to the antigen used in the vaccine. This allowed for comparison of the immune response (the number of T-cells that target a virus) in two different vaccine delivery methods.[15]
References
- ↑ "Tetramer Staining Guide". 2019. https://info.mblintl.com/tetramer-staining-guide-0-0.
- ↑ 2.0 2.1 2.2 "Major Histocompatibility Complex". InterPro. European Bioinformatics Institute (EMBL-EBI). https://www.ebi.ac.uk/interpro/potm/2005_2/Page2.htm.
- ↑ 3.0 3.1 "The Adaptive Immune Response: T lymphocytes and Their Functional Types". Anatomy and Physiology II. Lumen Learning. https://courses.lumenlearning.com/suny-ap2/chapter/the-adaptive-immune-response-t-lymphocytes-and-their-functional-types/.
- ↑ "MHC Tetramers". Medical and Biological Laboratories Co, Ltd (MBL). https://www.mblintl.com/products/mhc-tetramers/.
- ↑ 5.0 5.1 "MHC Tetramer Technology—Note 7.2". Thermo Fisher Scientific. https://www.thermofisher.com/us/en/home/references/molecular-probes-the-handbook/technical-notes-and-product-highlights/mhc-tetramer-technology.html.
- ↑ "Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996. 274: 94-96". Journal of Immunology 187 (1): 7–9. July 2011. PMID 21690331.
- ↑ Kimball, John W.. "Limiting Dilution Analysis". Kimball's Biology Pages. The Saylor Foundation. http://www.biology-pages.info/L/LimitingDilution.html.
- ↑ "A new look at T cells". The Journal of Experimental Medicine 187 (9): 1367–71. May 1998. doi:10.1084/jem.187.9.1367. PMID 9565629.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 "MHC-peptide tetramers for the analysis of antigen-specific T cells". Expert Review of Vaccines 9 (7): 765–74. July 2010. doi:10.1586/erv.10.66. PMID 20624049.
- ↑ "DIY - MHC class I Tetramers". 2019. https://immunaware.com/easymer-about/.
- ↑ "Staphylococcal phosphatidylglycerol antigens activate human T cells via CD1a". Nature Immunology 24 (1): 110–122. January 2023. doi:10.1038/s41590-022-01375-z. PMID 35265979.
- ↑ "Visualizing antigen specific CD4+ T cells using MHC class II tetramers". Journal of Visualized Experiments (25). March 2009. doi:10.3791/1167. PMID 19270641.
- ↑ "Invariant natural killer T cells: boon or bane in immunity to intracellular bacterial infections?" (in english). Journal of Innate Immunity 6 (5): 575–84. 2014. doi:10.1159/000361048. PMID 24903638.
- ↑ "Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA". Science 279 (5359): 2103–6. March 1998. doi:10.1126/science.279.5359.2103. PMID 9516110. Bibcode: 1998Sci...279.2103O. https://www.researchgate.net/publication/51312641.
- ↑ "+ T cell responses". Journal of Controlled Release 282: 120–130. July 2018. doi:10.1016/j.jconrel.2018.04.031. PMID 29673645.
Further reading
- "Phenotypic analysis of antigen-specific T lymphocytes". Science 274 (5284): 94–6. October 1996. doi:10.1126/science.274.5284.94. PMID 8810254. Bibcode: 1996Sci...274...94A.
 |