Medicine:Proliferative index
Proliferation, as one of the hallmarks and most fundamental biological processes in tumors,[1] is associated with tumor progression, response to therapy, and cancer patient survival.[2] Consequently, the evaluation of a tumor proliferative index (or growth fraction) has clinical significance in characterizing many solid tumors and hematologic malignancies.[3] This has led investigators to develop different technologies to evaluate the proliferation index in tumor samples. The most commonly used methods in evaluating a proliferative index include mitotic indexing, thymidine-labeling index, bromodeoxyuridine assay, the determination of fraction of cells in various phases of cell cycle, and the immunohistochemical evaluation of cell cycle-associated proteins.
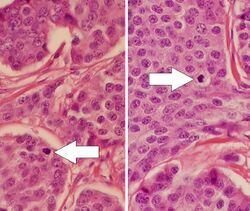
Mitotic index (also called mitotic count)
Mitotic indexing is the oldest method of assessing proliferation and is determined by counting the number of mitotic figures (cells undergoing mitosis) through a light microscope on H&E stained sections. It is usually expressed as the number of cells per microscopic field. Cells in the mitotic phase are identified by the typical appearance of their chromosomes in the cell during the mitotic phase of the cell cycle.[4] Usually the number of mitotic figures is expressed as the total number in a defined number of high power fields, such as 10 mitoses in 10 high power fields. Since the field of vision area can vary considerably between different microscopes, the exact area of the high power fields should be defined in order to compare results from different studies. Accordingly, one of the main problems of counting mitosis has been the reproducibility. Thus, the need for standardized methodology and strict protocols is important to achieve reproducible results.[5] Automated image alalysis using deep learning-based algorithms has been proposed as a promising tool to assist pathologists and thereby improve reproducibility and accuracy.[6]
Thymidine-labeling index
Thymidine-labeling indexing is determined by counting the number of tumor nuclei labeled on autoradiographed sections after incubating the tumor cells with thymidine. Rapidly proliferating cells readily update more radiolabeled Thymidine, which produces darker spots on the autoradiograph film.[7]
Bromodeoxyuridine assay
A bromodeoxyuridine assay, similar to thymidine-labeling indexing, incubates cells with radiolabeled bromodeoxyuridine, which is taken up at a greater rate by proliferating cells, and then uses a film to image the distribution of radiolabeled bromodeoxyuridine in cells.
S-phase fraction evaluation
Evaluating DNA histograms through flow cytometry provides an estimate of the fractions of cells within each of the phases in the cell cycle. Cell nuclei are stained with a DNA binding stain and the amount of staining is measured from the histogram.[8] The fractions of cells within the different cell cycle phases (G0/G1, S and G2/M compartments) can then be calculated from the histogram by computerized cell cycle analysis.
Immunohistochemical evaluation
The immunohistochemical detection of proliferation related proteins such as Ki-67 and proliferating cell nuclear antigen is a commonly used method to determine the proliferation index. Ki-67 is a nuclear antigen expressed in proliferating cells that is coded by the MKI67 gene on chromosome 10, and is expressed during the GI, S, G2, and M phases of the cell cycle. Cells are then stained with a Ki-67 antibody, and the number of stained nuclei is then expressed as a percentage of total tumor cells. It is recommended to count at least 500 tumor cells in the highest labeled area. The Ki-67 score closely correlates with other proliferation markers, and has been shown to have prognostic and predictive value for many different tumor types.[9] Similarly, proliferating cell nuclear antigen (PCNA) is a protein associated with cell proliferation that is upregulated in proliferating cells, making it another useful antigen for immunostaining.[10] It is associated with DNA polymerase alpha, which is expressed throughout the phases of the cell cycle. The expression of PCNA also correlates well with other proliferation markers such as mitotic count, S-phase fraction and Ki-67 staining.
Diagnostic role of proliferation index
The various methods of characterizing the proliferation index have found roles in both the diagnostic and prognostic evaluation of tumors. For instances, the number of mitotic cells is used to classify tumors. In general, a high proliferation index suggests malignancy and high-grade tumors.[11] Among solid tumors, the clinical significance of the proliferation index on breast cancer has been extensively studied. Mitotic counting has also been shown in multiple studies to have prognostic value in breast cancer, where a lower count of mitotic cells correlates with a more favorable outcome, and thus has been incorporated into part of the histological grading system. The Ki-67 labelling index has also been found to have prognostic significance where in many clinical practice guidelines, evaluation of Ki-67 in newly diagnosed invasive breast carcinomas is recommended.[12] Additionally, the tumor proliferation index has been used to predict the response to systemic chemotherapies in patients who are receiving neoadjuvant systemic therapy where patients who have tumors with high tumor proliferative index respond better to systemic cytotoxic therapies than those who have tumors with a low tumor proliferative index.[13]
References
- ↑ Hanahan, Douglas; Weinberg, Robert A. (2011). "Hallmarks of Cancer: The Next Generation". Cell 144 (5): 646–674. doi:10.1016/j.cell.2011.02.013. PMID 21376230.
- ↑ Yerushalmi, Rinat; Woods, Ryan; Ravdin, Peter M; Hayes, Malcolm M; Gelmon, Karen A (February 2010). "Ki67 in breast cancer: prognostic and predictive potential". The Lancet Oncology 11 (2): 174–183. doi:10.1016/s1470-2045(09)70262-1. PMID 20152769.
- ↑ Loo, Suet Kee; Ch'ng, Ewe Seng; Lawrie, Charles H.; Muruzabal, María Arestin; Gaafar, Ayman; Pomposo, María Puente; Husin, Azlan; Salleh, Md. Salzihan Md. et al. (December 2017). "DNMT1 is predictive of survival and associated with Ki-67 expression in R-CHOP-treated diffuse large B-cell lymphomas". Pathology 49 (7): 731–739. doi:10.1016/j.pathol.2017.08.009. PMID 29074044. https://ora.ox.ac.uk/objects/uuid:f5b74a28-1c4d-45f8-9d2f-64cb670d8fc8.
- ↑ Baak, J. P. (July 1990). "Mitosis counting in tumors". Human Pathology 21 (7): 683–685. doi:10.1016/0046-8177(90)90026-2. ISSN 0046-8177. PMID 2131787.
- ↑ van Diest, P. J.; Baak, J. P.; Matze-Cok, P.; Wisse-Brekelmans, E. C.; van Galen, C. M.; Kurver, P. H.; Bellot, S. M.; Fijnheer, J. et al. (June 1992). "Reproducibility of mitosis counting in 2,469 breast cancer specimens: results from the Multicenter Morphometric Mammary Carcinoma Project". Human Pathology 23 (6): 603–607. doi:10.1016/0046-8177(92)90313-r. ISSN 0046-8177. PMID 1592381.
- ↑ Bertram, Christof A; Aubreville, Marc; Donovan, Taryn A; Bartel, Alexander; Wilm, Frauke; Marzahl, Christian; Assenmacher, Charles-Antoine; Becker, Kathrin et al. (2021). "Computer-assisted mitotic count using a deep learning–based algorithm improves interobserver reproducibility and accuracy". Veterinary Pathology. doi:10.1177/03009858211067478. PMID 34965805.
- ↑ Silvestrini, R. (1994-10-01). "Cell kinetics: prognostic and therapeutic implications in human tumours" (in en). Cell Proliferation 27 (10): 579–596. doi:10.1111/j.1365-2184.1994.tb01376.x. ISSN 1365-2184.
- ↑ Wenger, C. R.; Beardslee, S.; Owens, M. A.; Pounds, G.; Oldaker, T.; Vendely, P.; Pandian, M. R.; Harrington, D. et al. (October 1993). "DNA ploidy, S-phase, and steroid receptors in more than 127,000 breast cancer patients". Breast Cancer Research and Treatment 28 (1): 9–20. doi:10.1007/BF00666351. ISSN 0167-6806. PMID 8123871.
- ↑ de Azambuja, E; Cardoso, F; Castro, G de; Colozza, M; Mano, M S; Durbecq, V; Sotiriou, C; Larsimont, D et al. (May 2007). "Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12 155 patients" (in En). British Journal of Cancer 96 (10): 1504–1513. doi:10.1038/sj.bjc.6603756. ISSN 1532-1827. PMID 17453008.
- ↑ Wang, Xing; Wang, Dong; Yuan, Na; Liu, Fanxiao; Wang, Fu; Wang, Bomin; Zhou, Dongsheng (2017-10-01). "The prognostic value of PCNA expression in patients with osteosarcoma" (in ENGLISH). Medicine 96 (41): e8254. doi:10.1097/md.0000000000008254. ISSN 0025-7974. PMID 29019895.
- ↑ Elston, C. W.; Ellis, I. O. (November 1991). "Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up". Histopathology 19 (5): 403–410. doi:10.1111/j.1365-2559.1991.tb00229.x. ISSN 0309-0167. PMID 1757079.
- ↑ Dowsett, Mitch; Nielsen, Torsten O.; A’Hern, Roger; Bartlett, John; Coombes, R. Charles; Cuzick, Jack; Ellis, Matthew; Henry, N. Lynn et al. (2011-11-16). "Assessment of Ki67 in Breast Cancer: Recommendations from the International Ki67 in Breast Cancer Working Group" (in en). JNCI Journal of the National Cancer Institute 103 (22): 1656–1664. doi:10.1093/jnci/djr393. ISSN 0027-8874. PMID 21960707.
- ↑ Groheux, David; Biard, L.; Lehmann-Che, J.; Teixeira, L.; Bouhidel, F. A.; Poirot, B.; Bertheau, P.; Merlet, P. et al. (2018-04-04). "Tumor metabolism assessed by FDG-PET/CT and tumor proliferation assessed by genomic grade index to predict response to neoadjuvant chemotherapy in triple negative breast cancer" (in en). European Journal of Nuclear Medicine and Molecular Imaging 45 (8): 1279–1288. doi:10.1007/s00259-018-3998-z. ISSN 1619-7070. PMID 29616304.
External links
- Proliferative index entry in the public domain NCI Dictionary of Cancer Terms
![]() This article incorporates public domain material from the U.S. National Cancer Institute document "Dictionary of Cancer Terms".
This article incorporates public domain material from the U.S. National Cancer Institute document "Dictionary of Cancer Terms".
 |