Chemistry:Bromodeoxyuridine
| |
| Names | |
|---|---|
| IUPAC name
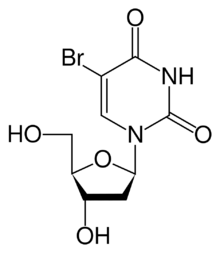
5-Bromo-2′-deoxyuridine
| |
| Systematic IUPAC name
5-Bromo-1-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidine-2,4(1H,3H)-dione | |
| Identifiers | |
3D model (JSmol)
|
|
| ChEMBL | |
| ChemSpider | |
| MeSH | Bromodeoxyuridine |
PubChem CID
|
|
| UNII | |
| |
| |
| Properties | |
| C9H11BrN2O5 | |
| Molar mass | 307.100 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Bromodeoxyuridine (5-bromo-2'-deoxyuridine, BrdU, BUdR, BrdUrd, broxuridine) is a synthetic nucleoside analogue with a chemical structure similar to thymidine. BrdU is commonly used to study cell proliferation in living tissues[1] and has been studied as a radiosensitizer[2] and diagnostic tool in people with cancer.[3]
During the S phase of the cell cycle (when DNA replication occurs), BrdU can be incorporated in place of thymidine in newly synthesized DNA molecules of dividing cells.[4] Cells that have recently performed DNA replication or DNA repair can be detected with antibodies specific for BrdU using techniques such as immunohistochemistry or immunofluorescence.[5] BrdU-labelled cells in humans can be detected up to two years after BrdU infusion.[6]
Because BrdU can replace thymidine during DNA replication, it can cause mutations, and its use is therefore potentially a health hazard.[citation needed] However, because it is neither radioactive nor myelotoxic at labeling concentrations, it is widely preferred for in vivo studies of cancer cell proliferation.[7][8] However, at radiosensitizing concentrations, BrdU becomes myelosuppressive, thus limiting its use for radiosensitizing.[2]
BrdU differs from thymidine in that BrdU substitutes a bromine atom for thymidine's CH3 group. The Br substitution can be used in X-ray diffraction experiments in crystals containing either DNA or RNA. The Br atom acts as an anomalous scatterer and its larger size will affect the crystal's X-ray diffraction enough to detect isomorphous differences as well.[9][10]
Bromodeoxyuridine releases gene silencing caused by DNA methylation.[11]
BrdU can also be used to identify microorganisms that respond to specific carbon substrates in aquatic[12] and soil[13] environments. A carbon substrate added to the incubations of environmental samples will cause the growth of microorganisms that can utilize that substrate. These microorganisms will then incorporate BrdU into their DNA as they grow. Community DNA can then be isolated and BrdU-labeled DNA purified using an immunocapture technique.[14] Subsequent sequencing of the labeled DNA can then be used to identify the microbial taxa that participated in the degradation of the added carbon source.
However, it is not certain whether all microbes present in an environmental sample can incorporate BrdU into their biomass during de novo DNA synthesis. Therefore, a group of microorganisms may respond to a carbon source but go undetected using this technique. Additionally, this technique is biased towards identifying microorganisms with A- and T-rich genomes.
DNA with BrdU transcribes as usual DNA, with guanine included in RNA as a complement to BrdU.[15]
See also
References
- ↑ Lehner, Bernadette; Sandner, Beatrice; Marschallinger, Julia; Lehner, Christine; Furtner, Tanja; Couillard-Despres, Sebastien; Rivera, Francisco J.; Brockhoff, Gero et al. (2011). "The dark side of BrdU in neural stem cell biology: Detrimental effects on cell cycle, differentiation and survival". Cell and Tissue Research 345 (3): 313–28. doi:10.1007/s00441-011-1213-7. PMID 21837406.
- ↑ 2.0 2.1 "Pharmacological evaluation of intravenous delivery of 5-bromodeoxyuridine to patients with brain tumors". Cancer Res. 44 (4): 1702–5. 1984. PMID 6704976.
- ↑ Dolbeare, F (May 1995). "Bromodeoxyuridine: a diagnostic tool in biology and medicine, Part I: Historical perspectives, histochemical methods and cell kinetics". The Histochemical Journal 27 (5): 339–69. doi:10.1007/BF02389022. PMID 7657555.
- ↑ Kee, N; S Sivalingam; R Boonstra; J.M Wojtowicz (March 2002). "The utility of Ki-67 and BrdU as proliferative markers of adult neurogenesis". Journal of Neuroscience Methods 115 (1): 97–105. doi:10.1016/S0165-0270(02)00007-9. PMID 11897369.
- ↑ Konishi, Teruaki; Takeyasu, Akihiro; Natsume, Toshiyuki; Furusawa, Yoshiya; Hieda, Kotaro (2011). "Visualization of Heavy Ion Tracks by Labeling 3'-OH Termini of Induced DNA Strand Breaks". Journal of Radiation Research 52 (4): 433–40. doi:10.1269/jrr.10097. PMID 21785232. Bibcode: 2011JRadR..52..433K.
- ↑ Eriksson, Peter; Ekaterina Perfilieva; Thomas Björk-Eriksson; Ann-Marie Alborn; Claes Nordborg; Daniel A. Peterson; Fred H. Gage (1998). "Neurogenesis in the adult human hippocampus". Nature Medicine. 1313-1317 4 (11): 1313–1317. doi:10.1038/3305. PMID 9809557.
- ↑ Fujimaki, Takamitsu; Masao Matsutani; Osamu Nakamura; Akio Asai; Nobuaki Funada; Morio Koike; Hiromu Segawa; Kouichi Aritake et al. (29 June 2006). "Correlation Between Bromodeoxyuridine- Labeling Indices and Patient Prognosis in Cerebral Astrocytic Tumors of Adults". Cancer 67 (6): 1629–1634. doi:10.1002/1097-0142(19910315)67:6<1629::AID-CNCR2820670626>3.0.CO;2-E. PMID 2001552.
- ↑ Hoshino, Takao; Tadashi Nagashima; Judith Murovic; Ellen M. Levin; Victor A. Levin; Stephen M. Rupp (1985). "Cell Kinetic Studies of In Situ Human Brain Tumors With Bromodeoxyuridine". Cytometry 6 (6): 627–632. doi:10.1002/cyto.990060619. PMID 2998714.
- ↑ Peterson, M. R.; Harrop, S. J.; McSweeney, S. M.; Leonard, G. A.; Thompson, A. W.; Hunter, W. N.; Helliwell, J. R. (1996). "MAD Phasing Strategies Explored with a Brominated Oligonucleotide Crystal at 1.65Å Resolution". Journal of Synchrotron Radiation 3 (Pt 1): 24–34. doi:10.1107/S0909049595013288. PMID 16702655.
- ↑ Beck, Tobias; Gruene, Tim; Sheldrick, George M. (2010). "The magic triangle goes MAD: Experimental phasing with a bromine derivative". Acta Crystallographica Section D 66 (4): 374–80. doi:10.1107/S0907444909051609. PMID 20382990.
- ↑ "On the concept and elucidation of endogenous retroviruses". Philos. Trans. R. Soc. Lond. B Biol. Sci. 368 (1626): 20120494. 2013. doi:10.1098/rstb.2012.0494. PMID 23938748.
- ↑ Tada, Yuya; Grossart, Hans-Peter (2013). "Community shifts of actively growing lake bacteria after N-acetyl-glucosamine addition: improving the BrdU-FACS method". The ISME Journal 8 (2): 441–454. doi:10.1038/ismej.2013.148. ISSN 1751-7362. PMID 23985742.
- ↑ "Culture-independent identification of microorganisms that respond to specified stimuli". Appl. Environ. Microbiol. 65 (8): 3398–400. 1999. doi:10.1128/AEM.65.8.3398-3400.1999. PMID 10427025. Bibcode: 1999ApEnM..65.3398B.
- ↑ Urbach, Ena; Kevin L. Vergin; Stephen J. Giovannoni (March 1999). "Immunochemical Detection and Isolation of DNA from Metabolically Active Bacteria". Applied and Environmental Microbiology 65 (3): 1207–1213. doi:10.1128/AEM.65.3.1207-1213.1999. PMID 10049885. Bibcode: 1999ApEnM..65.1207U.
- ↑ "Effect of 5-bromodeoxyuridine on the transcriptional properties of the genome in WI-38 human diploid fibroblasts". Chem. Biol. Interact. 10 (5): 363–375. 1975. doi:10.1016/0009-2797(75)90058-7. PMID 1095238.
External links
 |

